
Mycobacterium sp. 852013-51886_SCH5428379
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium; unclassified Mycobacterium
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
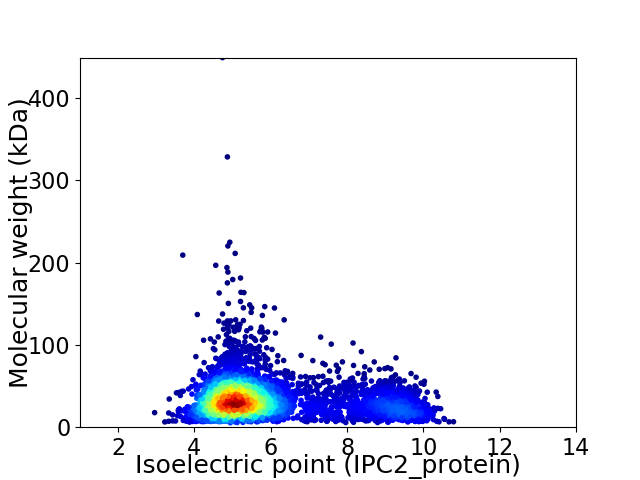
Virtual 2D-PAGE plot for 4393 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A0TCC2|A0A1A0TCC2_9MYCO Glycosyl transferase OS=Mycobacterium sp. 852013-51886_SCH5428379 OX=1834111 GN=A5757_03170 PE=4 SV=1
MM1 pKa = 7.73LLTLTACGSDD11 pKa = 4.38DD12 pKa = 3.49EE13 pKa = 4.67TAAAGPNSTVTPVASLAAAAVDD35 pKa = 4.14DD36 pKa = 4.43LQAYY40 pKa = 8.81WSEE43 pKa = 4.17QFPALYY49 pKa = 10.1GSEE52 pKa = 4.09YY53 pKa = 10.83VPLKK57 pKa = 10.2GGVYY61 pKa = 10.62ALTASSTKK69 pKa = 9.66WPEE72 pKa = 3.85CATEE76 pKa = 4.01YY77 pKa = 11.47ADD79 pKa = 4.03VEE81 pKa = 4.63GNAFFCQLDD90 pKa = 4.2DD91 pKa = 4.01SVSWDD96 pKa = 3.55AEE98 pKa = 4.0GLLPEE103 pKa = 4.59LQANFGDD110 pKa = 4.19FVIPVVLAHH119 pKa = 6.31EE120 pKa = 4.52WGHH123 pKa = 5.82AVQHH127 pKa = 6.33RR128 pKa = 11.84SGFFDD133 pKa = 3.62EE134 pKa = 4.77NALTVSSEE142 pKa = 3.96LQADD146 pKa = 4.58CFAGAWSRR154 pKa = 11.84HH155 pKa = 4.59ARR157 pKa = 11.84DD158 pKa = 2.96SGLFQVSDD166 pKa = 4.06DD167 pKa = 4.06QLDD170 pKa = 3.75DD171 pKa = 3.97ALAGVLDD178 pKa = 4.46LRR180 pKa = 11.84DD181 pKa = 3.74APGTSAEE188 pKa = 4.69DD189 pKa = 3.95PSAHH193 pKa = 6.18GSGFDD198 pKa = 3.73RR199 pKa = 11.84VAAFQDD205 pKa = 4.44GYY207 pKa = 11.41DD208 pKa = 3.73NAAGRR213 pKa = 11.84CKK215 pKa = 10.04EE216 pKa = 3.98YY217 pKa = 10.56RR218 pKa = 11.84DD219 pKa = 4.0GEE221 pKa = 4.36PTVLEE226 pKa = 4.69LPFEE230 pKa = 4.61DD231 pKa = 4.41AEE233 pKa = 4.36EE234 pKa = 4.13AASEE238 pKa = 4.34GNAPYY243 pKa = 10.74DD244 pKa = 3.79SIVNGVPYY252 pKa = 10.51DD253 pKa = 3.99LEE255 pKa = 5.44DD256 pKa = 3.69YY257 pKa = 7.47WTHH260 pKa = 6.06VYY262 pKa = 10.62PEE264 pKa = 4.07ITGGKK269 pKa = 7.89PWVPLRR275 pKa = 11.84GLEE278 pKa = 4.26PFDD281 pKa = 3.99PANPPMCDD289 pKa = 3.11GEE291 pKa = 6.07ADD293 pKa = 3.57DD294 pKa = 6.47DD295 pKa = 4.22YY296 pKa = 11.73TLFYY300 pKa = 10.5CVPDD304 pKa = 5.72DD305 pKa = 4.62FVAWDD310 pKa = 3.83NVEE313 pKa = 4.96DD314 pKa = 3.77MPEE317 pKa = 3.97VYY319 pKa = 10.04RR320 pKa = 11.84EE321 pKa = 4.17GGDD324 pKa = 3.33WAVGTLLATQYY335 pKa = 11.46GLAALTRR342 pKa = 11.84MGDD345 pKa = 3.29QSDD348 pKa = 4.23DD349 pKa = 3.47RR350 pKa = 11.84TSTLRR355 pKa = 11.84SDD357 pKa = 4.42CLAGSYY363 pKa = 7.66TASVMLFNRR372 pKa = 11.84ADD374 pKa = 3.39TSSFRR379 pKa = 11.84ISPGDD384 pKa = 3.54LDD386 pKa = 4.05EE387 pKa = 6.4GITALLVFRR396 pKa = 11.84GDD398 pKa = 3.21GDD400 pKa = 3.78VEE402 pKa = 4.28RR403 pKa = 11.84QGHH406 pKa = 5.1GFSRR410 pKa = 11.84VRR412 pKa = 11.84AYY414 pKa = 10.76RR415 pKa = 11.84EE416 pKa = 3.96GVLNGAEE423 pKa = 4.33GCLTYY428 pKa = 10.7QPP430 pKa = 4.68
MM1 pKa = 7.73LLTLTACGSDD11 pKa = 4.38DD12 pKa = 3.49EE13 pKa = 4.67TAAAGPNSTVTPVASLAAAAVDD35 pKa = 4.14DD36 pKa = 4.43LQAYY40 pKa = 8.81WSEE43 pKa = 4.17QFPALYY49 pKa = 10.1GSEE52 pKa = 4.09YY53 pKa = 10.83VPLKK57 pKa = 10.2GGVYY61 pKa = 10.62ALTASSTKK69 pKa = 9.66WPEE72 pKa = 3.85CATEE76 pKa = 4.01YY77 pKa = 11.47ADD79 pKa = 4.03VEE81 pKa = 4.63GNAFFCQLDD90 pKa = 4.2DD91 pKa = 4.01SVSWDD96 pKa = 3.55AEE98 pKa = 4.0GLLPEE103 pKa = 4.59LQANFGDD110 pKa = 4.19FVIPVVLAHH119 pKa = 6.31EE120 pKa = 4.52WGHH123 pKa = 5.82AVQHH127 pKa = 6.33RR128 pKa = 11.84SGFFDD133 pKa = 3.62EE134 pKa = 4.77NALTVSSEE142 pKa = 3.96LQADD146 pKa = 4.58CFAGAWSRR154 pKa = 11.84HH155 pKa = 4.59ARR157 pKa = 11.84DD158 pKa = 2.96SGLFQVSDD166 pKa = 4.06DD167 pKa = 4.06QLDD170 pKa = 3.75DD171 pKa = 3.97ALAGVLDD178 pKa = 4.46LRR180 pKa = 11.84DD181 pKa = 3.74APGTSAEE188 pKa = 4.69DD189 pKa = 3.95PSAHH193 pKa = 6.18GSGFDD198 pKa = 3.73RR199 pKa = 11.84VAAFQDD205 pKa = 4.44GYY207 pKa = 11.41DD208 pKa = 3.73NAAGRR213 pKa = 11.84CKK215 pKa = 10.04EE216 pKa = 3.98YY217 pKa = 10.56RR218 pKa = 11.84DD219 pKa = 4.0GEE221 pKa = 4.36PTVLEE226 pKa = 4.69LPFEE230 pKa = 4.61DD231 pKa = 4.41AEE233 pKa = 4.36EE234 pKa = 4.13AASEE238 pKa = 4.34GNAPYY243 pKa = 10.74DD244 pKa = 3.79SIVNGVPYY252 pKa = 10.51DD253 pKa = 3.99LEE255 pKa = 5.44DD256 pKa = 3.69YY257 pKa = 7.47WTHH260 pKa = 6.06VYY262 pKa = 10.62PEE264 pKa = 4.07ITGGKK269 pKa = 7.89PWVPLRR275 pKa = 11.84GLEE278 pKa = 4.26PFDD281 pKa = 3.99PANPPMCDD289 pKa = 3.11GEE291 pKa = 6.07ADD293 pKa = 3.57DD294 pKa = 6.47DD295 pKa = 4.22YY296 pKa = 11.73TLFYY300 pKa = 10.5CVPDD304 pKa = 5.72DD305 pKa = 4.62FVAWDD310 pKa = 3.83NVEE313 pKa = 4.96DD314 pKa = 3.77MPEE317 pKa = 3.97VYY319 pKa = 10.04RR320 pKa = 11.84EE321 pKa = 4.17GGDD324 pKa = 3.33WAVGTLLATQYY335 pKa = 11.46GLAALTRR342 pKa = 11.84MGDD345 pKa = 3.29QSDD348 pKa = 4.23DD349 pKa = 3.47RR350 pKa = 11.84TSTLRR355 pKa = 11.84SDD357 pKa = 4.42CLAGSYY363 pKa = 7.66TASVMLFNRR372 pKa = 11.84ADD374 pKa = 3.39TSSFRR379 pKa = 11.84ISPGDD384 pKa = 3.54LDD386 pKa = 4.05EE387 pKa = 6.4GITALLVFRR396 pKa = 11.84GDD398 pKa = 3.21GDD400 pKa = 3.78VEE402 pKa = 4.28RR403 pKa = 11.84QGHH406 pKa = 5.1GFSRR410 pKa = 11.84VRR412 pKa = 11.84AYY414 pKa = 10.76RR415 pKa = 11.84EE416 pKa = 3.96GVLNGAEE423 pKa = 4.33GCLTYY428 pKa = 10.7QPP430 pKa = 4.68
Molecular weight: 46.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A0TBW8|A0A1A0TBW8_9MYCO Xaa-Pro aminopeptidase OS=Mycobacterium sp. 852013-51886_SCH5428379 OX=1834111 GN=A5757_02230 PE=4 SV=1
MM1 pKa = 7.81ASTPNMSTPPVMIAIDD17 pKa = 3.4PHH19 pKa = 6.46KK20 pKa = 10.61RR21 pKa = 11.84SWTAAAVDD29 pKa = 4.01VRR31 pKa = 11.84LQPLATIRR39 pKa = 11.84VPVSRR44 pKa = 11.84DD45 pKa = 3.0GYY47 pKa = 8.86RR48 pKa = 11.84TLLRR52 pKa = 11.84FARR55 pKa = 11.84RR56 pKa = 11.84WPDD59 pKa = 3.25TAWAIEE65 pKa = 4.17GASGLGAPLTVRR77 pKa = 11.84LRR79 pKa = 11.84QDD81 pKa = 4.14GITVLDD87 pKa = 4.21VPAKK91 pKa = 9.35LAARR95 pKa = 11.84VRR97 pKa = 11.84LLSTGHH103 pKa = 5.72GRR105 pKa = 11.84KK106 pKa = 9.21SDD108 pKa = 3.47EE109 pKa = 3.97ADD111 pKa = 3.19AVSVGVAAHH120 pKa = 6.41TSSSLNTAEE129 pKa = 4.66IDD131 pKa = 3.74SATTALRR138 pKa = 11.84ALVEE142 pKa = 4.15HH143 pKa = 7.25RR144 pKa = 11.84DD145 pKa = 3.86DD146 pKa = 3.86LVKK149 pKa = 10.26TRR151 pKa = 11.84TQTINRR157 pKa = 11.84LHH159 pKa = 6.11TVLTHH164 pKa = 6.8LIPGGASRR172 pKa = 11.84DD173 pKa = 3.81LNAEE177 pKa = 3.78RR178 pKa = 11.84AAEE181 pKa = 4.03LLRR184 pKa = 11.84TVRR187 pKa = 11.84PRR189 pKa = 11.84DD190 pKa = 3.46TAGKK194 pKa = 7.11TLRR197 pKa = 11.84ALAADD202 pKa = 5.57LISEE206 pKa = 4.21LRR208 pKa = 11.84GLDD211 pKa = 3.28RR212 pKa = 11.84RR213 pKa = 11.84IDD215 pKa = 3.7KK216 pKa = 10.39AVHH219 pKa = 6.62DD220 pKa = 4.23IEE222 pKa = 5.55IAVKK226 pKa = 10.65DD227 pKa = 3.87CGTTLTEE234 pKa = 3.77LHH236 pKa = 6.89GIGALTAAKK245 pKa = 9.44IVSRR249 pKa = 11.84VGSIQRR255 pKa = 11.84FDD257 pKa = 3.52SPAAFASYY265 pKa = 9.87TGTAPIEE272 pKa = 4.21VSSGDD277 pKa = 3.71VVRR280 pKa = 11.84HH281 pKa = 5.25RR282 pKa = 11.84LSRR285 pKa = 11.84AGDD288 pKa = 3.46RR289 pKa = 11.84QLNYY293 pKa = 9.25CLHH296 pKa = 6.66VMAITQIRR304 pKa = 11.84RR305 pKa = 11.84DD306 pKa = 3.82TPGRR310 pKa = 11.84TYY312 pKa = 11.11YY313 pKa = 10.33LRR315 pKa = 11.84KK316 pKa = 9.02RR317 pKa = 11.84ANGKK321 pKa = 8.19SHH323 pKa = 6.54SEE325 pKa = 3.53AMRR328 pKa = 11.84CLKK331 pKa = 10.35RR332 pKa = 11.84RR333 pKa = 11.84LSDD336 pKa = 3.16TVYY339 pKa = 10.67RR340 pKa = 11.84QLLRR344 pKa = 11.84DD345 pKa = 3.73ANTTTT350 pKa = 3.72
MM1 pKa = 7.81ASTPNMSTPPVMIAIDD17 pKa = 3.4PHH19 pKa = 6.46KK20 pKa = 10.61RR21 pKa = 11.84SWTAAAVDD29 pKa = 4.01VRR31 pKa = 11.84LQPLATIRR39 pKa = 11.84VPVSRR44 pKa = 11.84DD45 pKa = 3.0GYY47 pKa = 8.86RR48 pKa = 11.84TLLRR52 pKa = 11.84FARR55 pKa = 11.84RR56 pKa = 11.84WPDD59 pKa = 3.25TAWAIEE65 pKa = 4.17GASGLGAPLTVRR77 pKa = 11.84LRR79 pKa = 11.84QDD81 pKa = 4.14GITVLDD87 pKa = 4.21VPAKK91 pKa = 9.35LAARR95 pKa = 11.84VRR97 pKa = 11.84LLSTGHH103 pKa = 5.72GRR105 pKa = 11.84KK106 pKa = 9.21SDD108 pKa = 3.47EE109 pKa = 3.97ADD111 pKa = 3.19AVSVGVAAHH120 pKa = 6.41TSSSLNTAEE129 pKa = 4.66IDD131 pKa = 3.74SATTALRR138 pKa = 11.84ALVEE142 pKa = 4.15HH143 pKa = 7.25RR144 pKa = 11.84DD145 pKa = 3.86DD146 pKa = 3.86LVKK149 pKa = 10.26TRR151 pKa = 11.84TQTINRR157 pKa = 11.84LHH159 pKa = 6.11TVLTHH164 pKa = 6.8LIPGGASRR172 pKa = 11.84DD173 pKa = 3.81LNAEE177 pKa = 3.78RR178 pKa = 11.84AAEE181 pKa = 4.03LLRR184 pKa = 11.84TVRR187 pKa = 11.84PRR189 pKa = 11.84DD190 pKa = 3.46TAGKK194 pKa = 7.11TLRR197 pKa = 11.84ALAADD202 pKa = 5.57LISEE206 pKa = 4.21LRR208 pKa = 11.84GLDD211 pKa = 3.28RR212 pKa = 11.84RR213 pKa = 11.84IDD215 pKa = 3.7KK216 pKa = 10.39AVHH219 pKa = 6.62DD220 pKa = 4.23IEE222 pKa = 5.55IAVKK226 pKa = 10.65DD227 pKa = 3.87CGTTLTEE234 pKa = 3.77LHH236 pKa = 6.89GIGALTAAKK245 pKa = 9.44IVSRR249 pKa = 11.84VGSIQRR255 pKa = 11.84FDD257 pKa = 3.52SPAAFASYY265 pKa = 9.87TGTAPIEE272 pKa = 4.21VSSGDD277 pKa = 3.71VVRR280 pKa = 11.84HH281 pKa = 5.25RR282 pKa = 11.84LSRR285 pKa = 11.84AGDD288 pKa = 3.46RR289 pKa = 11.84QLNYY293 pKa = 9.25CLHH296 pKa = 6.66VMAITQIRR304 pKa = 11.84RR305 pKa = 11.84DD306 pKa = 3.82TPGRR310 pKa = 11.84TYY312 pKa = 11.11YY313 pKa = 10.33LRR315 pKa = 11.84KK316 pKa = 9.02RR317 pKa = 11.84ANGKK321 pKa = 8.19SHH323 pKa = 6.54SEE325 pKa = 3.53AMRR328 pKa = 11.84CLKK331 pKa = 10.35RR332 pKa = 11.84RR333 pKa = 11.84LSDD336 pKa = 3.16TVYY339 pKa = 10.67RR340 pKa = 11.84QLLRR344 pKa = 11.84DD345 pKa = 3.73ANTTTT350 pKa = 3.72
Molecular weight: 38.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1448496 |
48 |
4179 |
329.7 |
35.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.28 ± 0.051 | 0.766 ± 0.009 |
6.529 ± 0.036 | 5.285 ± 0.034 |
3.026 ± 0.02 | 9.088 ± 0.094 |
2.139 ± 0.018 | 4.135 ± 0.024 |
1.962 ± 0.029 | 9.834 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.036 ± 0.016 | 2.082 ± 0.022 |
5.834 ± 0.037 | 2.852 ± 0.02 |
7.415 ± 0.041 | 5.159 ± 0.025 |
6.115 ± 0.03 | 8.894 ± 0.032 |
1.482 ± 0.016 | 2.086 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
