
Wuhan insect virus 26
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
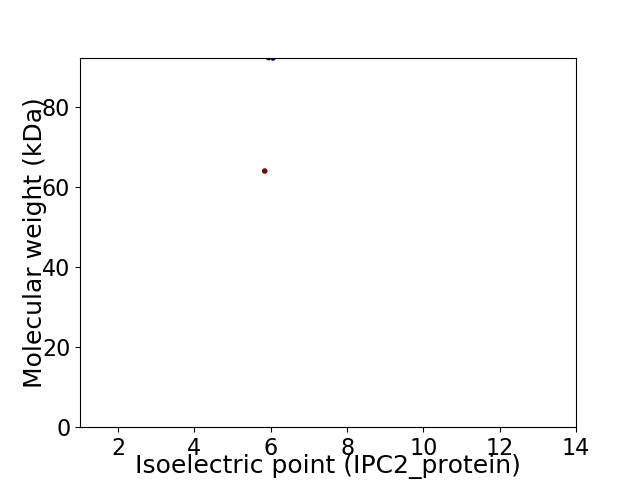
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFB4|A0A1L3KFB4_9VIRU Uncharacterized protein OS=Wuhan insect virus 26 OX=1923730 PE=4 SV=1
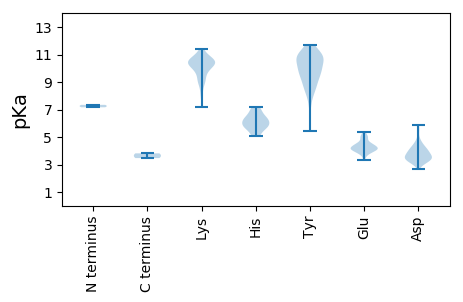
MM1 pKa = 7.16LASWLSARR9 pKa = 11.84LYY11 pKa = 10.67RR12 pKa = 11.84DD13 pKa = 3.35NKK15 pKa = 10.08DD16 pKa = 2.64TDD18 pKa = 3.51NILRR22 pKa = 11.84VKK24 pKa = 9.53TSPYY28 pKa = 9.72SDD30 pKa = 3.27SHH32 pKa = 6.63VICPLDD38 pKa = 4.2RR39 pKa = 11.84EE40 pKa = 4.69AEE42 pKa = 4.14DD43 pKa = 4.64HH44 pKa = 5.66IHH46 pKa = 6.62EE47 pKa = 5.1IEE49 pKa = 4.87LGPPVDD55 pKa = 3.57QEE57 pKa = 4.85LINGATWMLRR67 pKa = 11.84AKK69 pKa = 10.42DD70 pKa = 4.09SYY72 pKa = 10.09WNRR75 pKa = 11.84PYY77 pKa = 10.96VLHH80 pKa = 7.13INGATTGQVEE90 pKa = 5.34FYY92 pKa = 10.19LSHH95 pKa = 6.0VLGRR99 pKa = 11.84KK100 pKa = 8.93RR101 pKa = 11.84RR102 pKa = 11.84SALNYY107 pKa = 9.55DD108 pKa = 3.85VEE110 pKa = 4.59IEE112 pKa = 4.16GLNSRR117 pKa = 11.84EE118 pKa = 4.08LYY120 pKa = 10.8LDD122 pKa = 3.19IVNGDD127 pKa = 3.2GSEE130 pKa = 4.08PGWFHH135 pKa = 7.07EE136 pKa = 5.02IPWTSPDD143 pKa = 4.04ILWSWTLDD151 pKa = 3.55YY152 pKa = 11.6VKK154 pKa = 10.8LNRR157 pKa = 11.84LEE159 pKa = 3.98QAFAAAFEE167 pKa = 4.24TLAAVAIQPQWSSLEE182 pKa = 3.73ACAWQSSQIVVSFGAFSPTRR202 pKa = 11.84ARR204 pKa = 11.84IRR206 pKa = 11.84SNLTGTPYY214 pKa = 10.68TPFAEE219 pKa = 4.76ASSFMLDD226 pKa = 3.13EE227 pKa = 4.58AAAPTHH233 pKa = 6.27YY234 pKa = 8.98FTAAAIANYY243 pKa = 8.54YY244 pKa = 7.88MWYY247 pKa = 10.64GLYY250 pKa = 9.81TLLHH254 pKa = 5.95NEE256 pKa = 3.32ARR258 pKa = 11.84TRR260 pKa = 11.84NSWKK264 pKa = 9.13TVFSSVADD272 pKa = 4.11EE273 pKa = 5.11LKK275 pKa = 10.28MLYY278 pKa = 9.64TPMMRR283 pKa = 11.84AACVSVITGRR293 pKa = 11.84EE294 pKa = 3.72YY295 pKa = 11.02ASCMNHH301 pKa = 5.87GAGMYY306 pKa = 9.71IDD308 pKa = 4.04MSSMYY313 pKa = 8.24KK314 pKa = 9.62QKK316 pKa = 11.07YY317 pKa = 8.71IGPLVPLDD325 pKa = 3.72GTVGTSVEE333 pKa = 4.02IEE335 pKa = 4.34AIYY338 pKa = 10.82APVSGGLVLGTMTGEE353 pKa = 4.51FEE355 pKa = 4.41TTAHH359 pKa = 5.67LTGVRR364 pKa = 11.84ALEE367 pKa = 4.32CKK369 pKa = 10.44GVVGKK374 pKa = 10.61AFDD377 pKa = 4.01LKK379 pKa = 10.53EE380 pKa = 4.13LSVLATMYY388 pKa = 10.96RR389 pKa = 11.84LFGYY393 pKa = 9.53EE394 pKa = 5.33LEE396 pKa = 4.56LKK398 pKa = 10.25DD399 pKa = 4.04YY400 pKa = 8.69QTGIIKK406 pKa = 8.52RR407 pKa = 11.84TWAPVRR413 pKa = 11.84DD414 pKa = 4.22CVVDD418 pKa = 3.58PATIEE423 pKa = 3.91FDD425 pKa = 4.51PISPRR430 pKa = 11.84LWMIEE435 pKa = 3.77GAQPRR440 pKa = 11.84EE441 pKa = 4.26GRR443 pKa = 11.84RR444 pKa = 11.84HH445 pKa = 7.18IIPTPQNLLSGEE457 pKa = 4.14PVVIVHH463 pKa = 6.17QKK465 pKa = 8.11PTISVTKK472 pKa = 8.89WKK474 pKa = 10.76SRR476 pKa = 11.84TKK478 pKa = 10.21PPRR481 pKa = 11.84PFIATSRR488 pKa = 11.84VKK490 pKa = 10.96APVQFLIKK498 pKa = 10.58APFEE502 pKa = 4.11YY503 pKa = 10.91NPISFEE509 pKa = 4.04AKK511 pKa = 8.98EE512 pKa = 4.27VPSHH516 pKa = 6.4KK517 pKa = 10.33FKK519 pKa = 11.26LSDD522 pKa = 3.58FQEE525 pKa = 4.98GKK527 pKa = 9.88SQVPPKK533 pKa = 10.33KK534 pKa = 10.22PEE536 pKa = 3.78GQKK539 pKa = 9.95ILNQRR544 pKa = 11.84TAEE547 pKa = 4.04PARR550 pKa = 11.84EE551 pKa = 4.01EE552 pKa = 4.08EE553 pKa = 4.6SIPTAGAAASLEE565 pKa = 4.28SSLQQQ570 pKa = 3.47
MM1 pKa = 7.16LASWLSARR9 pKa = 11.84LYY11 pKa = 10.67RR12 pKa = 11.84DD13 pKa = 3.35NKK15 pKa = 10.08DD16 pKa = 2.64TDD18 pKa = 3.51NILRR22 pKa = 11.84VKK24 pKa = 9.53TSPYY28 pKa = 9.72SDD30 pKa = 3.27SHH32 pKa = 6.63VICPLDD38 pKa = 4.2RR39 pKa = 11.84EE40 pKa = 4.69AEE42 pKa = 4.14DD43 pKa = 4.64HH44 pKa = 5.66IHH46 pKa = 6.62EE47 pKa = 5.1IEE49 pKa = 4.87LGPPVDD55 pKa = 3.57QEE57 pKa = 4.85LINGATWMLRR67 pKa = 11.84AKK69 pKa = 10.42DD70 pKa = 4.09SYY72 pKa = 10.09WNRR75 pKa = 11.84PYY77 pKa = 10.96VLHH80 pKa = 7.13INGATTGQVEE90 pKa = 5.34FYY92 pKa = 10.19LSHH95 pKa = 6.0VLGRR99 pKa = 11.84KK100 pKa = 8.93RR101 pKa = 11.84RR102 pKa = 11.84SALNYY107 pKa = 9.55DD108 pKa = 3.85VEE110 pKa = 4.59IEE112 pKa = 4.16GLNSRR117 pKa = 11.84EE118 pKa = 4.08LYY120 pKa = 10.8LDD122 pKa = 3.19IVNGDD127 pKa = 3.2GSEE130 pKa = 4.08PGWFHH135 pKa = 7.07EE136 pKa = 5.02IPWTSPDD143 pKa = 4.04ILWSWTLDD151 pKa = 3.55YY152 pKa = 11.6VKK154 pKa = 10.8LNRR157 pKa = 11.84LEE159 pKa = 3.98QAFAAAFEE167 pKa = 4.24TLAAVAIQPQWSSLEE182 pKa = 3.73ACAWQSSQIVVSFGAFSPTRR202 pKa = 11.84ARR204 pKa = 11.84IRR206 pKa = 11.84SNLTGTPYY214 pKa = 10.68TPFAEE219 pKa = 4.76ASSFMLDD226 pKa = 3.13EE227 pKa = 4.58AAAPTHH233 pKa = 6.27YY234 pKa = 8.98FTAAAIANYY243 pKa = 8.54YY244 pKa = 7.88MWYY247 pKa = 10.64GLYY250 pKa = 9.81TLLHH254 pKa = 5.95NEE256 pKa = 3.32ARR258 pKa = 11.84TRR260 pKa = 11.84NSWKK264 pKa = 9.13TVFSSVADD272 pKa = 4.11EE273 pKa = 5.11LKK275 pKa = 10.28MLYY278 pKa = 9.64TPMMRR283 pKa = 11.84AACVSVITGRR293 pKa = 11.84EE294 pKa = 3.72YY295 pKa = 11.02ASCMNHH301 pKa = 5.87GAGMYY306 pKa = 9.71IDD308 pKa = 4.04MSSMYY313 pKa = 8.24KK314 pKa = 9.62QKK316 pKa = 11.07YY317 pKa = 8.71IGPLVPLDD325 pKa = 3.72GTVGTSVEE333 pKa = 4.02IEE335 pKa = 4.34AIYY338 pKa = 10.82APVSGGLVLGTMTGEE353 pKa = 4.51FEE355 pKa = 4.41TTAHH359 pKa = 5.67LTGVRR364 pKa = 11.84ALEE367 pKa = 4.32CKK369 pKa = 10.44GVVGKK374 pKa = 10.61AFDD377 pKa = 4.01LKK379 pKa = 10.53EE380 pKa = 4.13LSVLATMYY388 pKa = 10.96RR389 pKa = 11.84LFGYY393 pKa = 9.53EE394 pKa = 5.33LEE396 pKa = 4.56LKK398 pKa = 10.25DD399 pKa = 4.04YY400 pKa = 8.69QTGIIKK406 pKa = 8.52RR407 pKa = 11.84TWAPVRR413 pKa = 11.84DD414 pKa = 4.22CVVDD418 pKa = 3.58PATIEE423 pKa = 3.91FDD425 pKa = 4.51PISPRR430 pKa = 11.84LWMIEE435 pKa = 3.77GAQPRR440 pKa = 11.84EE441 pKa = 4.26GRR443 pKa = 11.84RR444 pKa = 11.84HH445 pKa = 7.18IIPTPQNLLSGEE457 pKa = 4.14PVVIVHH463 pKa = 6.17QKK465 pKa = 8.11PTISVTKK472 pKa = 8.89WKK474 pKa = 10.76SRR476 pKa = 11.84TKK478 pKa = 10.21PPRR481 pKa = 11.84PFIATSRR488 pKa = 11.84VKK490 pKa = 10.96APVQFLIKK498 pKa = 10.58APFEE502 pKa = 4.11YY503 pKa = 10.91NPISFEE509 pKa = 4.04AKK511 pKa = 8.98EE512 pKa = 4.27VPSHH516 pKa = 6.4KK517 pKa = 10.33FKK519 pKa = 11.26LSDD522 pKa = 3.58FQEE525 pKa = 4.98GKK527 pKa = 9.88SQVPPKK533 pKa = 10.33KK534 pKa = 10.22PEE536 pKa = 3.78GQKK539 pKa = 9.95ILNQRR544 pKa = 11.84TAEE547 pKa = 4.04PARR550 pKa = 11.84EE551 pKa = 4.01EE552 pKa = 4.08EE553 pKa = 4.6SIPTAGAAASLEE565 pKa = 4.28SSLQQQ570 pKa = 3.47
Molecular weight: 63.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFB4|A0A1L3KFB4_9VIRU Uncharacterized protein OS=Wuhan insect virus 26 OX=1923730 PE=4 SV=1
MM1 pKa = 7.34QEE3 pKa = 4.09RR4 pKa = 11.84DD5 pKa = 3.46SFTALDD11 pKa = 3.35ICDD14 pKa = 3.35GMRR17 pKa = 11.84EE18 pKa = 4.25GRR20 pKa = 11.84EE21 pKa = 3.51HH22 pKa = 5.93TFAVRR27 pKa = 11.84SGHH30 pKa = 5.73VPTGLRR36 pKa = 11.84ITCNGTYY43 pKa = 10.21LGVPFKK49 pKa = 10.56QAEE52 pKa = 4.21LLLIDD57 pKa = 4.57RR58 pKa = 11.84LPAKK62 pKa = 10.39LHH64 pKa = 5.7EE65 pKa = 5.19CNVAYY70 pKa = 10.06DD71 pKa = 3.63YY72 pKa = 11.65YY73 pKa = 9.94GTVVSARR80 pKa = 11.84LVPGRR85 pKa = 11.84DD86 pKa = 3.03ATYY89 pKa = 9.5VYY91 pKa = 10.72YY92 pKa = 10.88YY93 pKa = 10.38VDD95 pKa = 3.48QDD97 pKa = 3.76IQPVTQNLLGMLSRR111 pKa = 11.84HH112 pKa = 5.56FLGDD116 pKa = 3.14FSGYY120 pKa = 9.9YY121 pKa = 10.14NDD123 pKa = 3.8WCSLDD128 pKa = 3.39NVFYY132 pKa = 11.11SLSKK136 pKa = 10.45CDD138 pKa = 3.82NPVKK142 pKa = 10.39RR143 pKa = 11.84HH144 pKa = 5.12TIKK147 pKa = 10.22TIRR150 pKa = 11.84DD151 pKa = 3.42LPKK154 pKa = 9.78PKK156 pKa = 9.84ISGAHH161 pKa = 6.42HH162 pKa = 5.9IHH164 pKa = 5.41YY165 pKa = 7.07TASEE169 pKa = 4.26VWSVLDD175 pKa = 5.52DD176 pKa = 3.95RR177 pKa = 11.84GKK179 pKa = 10.59DD180 pKa = 3.43AARR183 pKa = 11.84HH184 pKa = 5.05ALRR187 pKa = 11.84LPADD191 pKa = 3.54ATTSFVGGVMLWLASLPTEE210 pKa = 4.15LLNPIIEE217 pKa = 4.53SDD219 pKa = 3.79LLDD222 pKa = 4.6AEE224 pKa = 4.45DD225 pKa = 4.19TIEE228 pKa = 4.36FGKK231 pKa = 10.36KK232 pKa = 9.3AKK234 pKa = 9.99KK235 pKa = 10.42LSVTAKK241 pKa = 9.18SFQNIVEE248 pKa = 4.23ADD250 pKa = 3.44LRR252 pKa = 11.84PIFEE256 pKa = 5.0ADD258 pKa = 3.32VLVNRR263 pKa = 11.84DD264 pKa = 3.22VGEE267 pKa = 4.06VDD269 pKa = 2.96WAGEE273 pKa = 4.0KK274 pKa = 10.67NNRR277 pKa = 11.84CKK279 pKa = 10.63PNLASVSPGEE289 pKa = 4.34VYY291 pKa = 10.91ARR293 pKa = 11.84TLALFTKK300 pKa = 10.34EE301 pKa = 4.44DD302 pKa = 3.95DD303 pKa = 3.92EE304 pKa = 4.6KK305 pKa = 11.4ALPRR309 pKa = 11.84RR310 pKa = 11.84LEE312 pKa = 3.92WEE314 pKa = 4.38KK315 pKa = 10.87FWDD318 pKa = 5.53ARR320 pKa = 11.84WQWSASGSIHH330 pKa = 6.31SQYY333 pKa = 10.91QEE335 pKa = 4.93DD336 pKa = 4.84LDD338 pKa = 4.13SLPKK342 pKa = 10.32EE343 pKa = 3.91RR344 pKa = 11.84EE345 pKa = 4.05LKK347 pKa = 10.82NKK349 pKa = 10.1FIALSMQGDD358 pKa = 3.82MPFEE362 pKa = 4.47HH363 pKa = 6.8FLNRR367 pKa = 11.84KK368 pKa = 8.12PEE370 pKa = 3.8IVAWSSVKK378 pKa = 10.74YY379 pKa = 9.63EE380 pKa = 3.89WGKK383 pKa = 9.08MRR385 pKa = 11.84AIYY388 pKa = 8.58GTDD391 pKa = 2.91LTSYY395 pKa = 10.79VLAHH399 pKa = 5.71YY400 pKa = 10.98AFFNCEE406 pKa = 3.81DD407 pKa = 4.08TLPTDD412 pKa = 4.19FPVGEE417 pKa = 4.21KK418 pKa = 10.28ARR420 pKa = 11.84PSFVSARR427 pKa = 11.84VAAILEE433 pKa = 4.51GTVPLCVDD441 pKa = 4.59FEE443 pKa = 4.98DD444 pKa = 5.9FNSQHH449 pKa = 6.83SNDD452 pKa = 3.2AMEE455 pKa = 5.1AVVQAYY461 pKa = 9.25IDD463 pKa = 4.37AYY465 pKa = 8.43SQYY468 pKa = 7.75MTPEE472 pKa = 4.02QVQAAMWTRR481 pKa = 11.84EE482 pKa = 4.39SISNTRR488 pKa = 11.84VIDD491 pKa = 3.62NMGTKK496 pKa = 9.69TSYY499 pKa = 8.55KK500 pKa = 9.92TKK502 pKa = 8.99GTLMSGWRR510 pKa = 11.84LTTFINSVLNYY521 pKa = 9.94VYY523 pKa = 9.67TQALISGSHH532 pKa = 5.12AMSRR536 pKa = 11.84SVHH539 pKa = 6.05NGDD542 pKa = 5.05DD543 pKa = 3.48VLLGIRR549 pKa = 11.84NFKK552 pKa = 9.78IVRR555 pKa = 11.84DD556 pKa = 3.79IVARR560 pKa = 11.84ADD562 pKa = 3.4KK563 pKa = 11.23YY564 pKa = 10.77NVRR567 pKa = 11.84LQRR570 pKa = 11.84TKK572 pKa = 10.81CAFGGLAEE580 pKa = 4.41FLRR583 pKa = 11.84VDD585 pKa = 4.33HH586 pKa = 6.34MRR588 pKa = 11.84GDD590 pKa = 3.57YY591 pKa = 10.53GQYY594 pKa = 8.72LTRR597 pKa = 11.84NIATVMHH604 pKa = 6.26SRR606 pKa = 11.84IEE608 pKa = 4.14SKK610 pKa = 10.49IAVSAVDD617 pKa = 3.43VVQAMEE623 pKa = 4.21DD624 pKa = 3.19RR625 pKa = 11.84LRR627 pKa = 11.84EE628 pKa = 3.81YY629 pKa = 10.48LQRR632 pKa = 11.84GGVWDD637 pKa = 3.61YY638 pKa = 11.72AVQLRR643 pKa = 11.84EE644 pKa = 3.98LYY646 pKa = 9.17YY647 pKa = 11.19DD648 pKa = 3.88RR649 pKa = 11.84MAPIYY654 pKa = 9.65SLRR657 pKa = 11.84PQDD660 pKa = 4.35LYY662 pKa = 11.27DD663 pKa = 3.14IRR665 pKa = 11.84TAHH668 pKa = 6.22RR669 pKa = 11.84VVGGIAEE676 pKa = 4.24DD677 pKa = 3.54VSARR681 pKa = 11.84VDD683 pKa = 3.23KK684 pKa = 10.52FIRR687 pKa = 11.84TSSEE691 pKa = 4.14SIEE694 pKa = 4.27TTLPDD699 pKa = 3.74TLPGVDD705 pKa = 3.73AYY707 pKa = 10.94ARR709 pKa = 11.84LLKK712 pKa = 10.36KK713 pKa = 10.55VLEE716 pKa = 4.35LEE718 pKa = 4.49VPVSVVSTRR727 pKa = 11.84IRR729 pKa = 11.84NATLNAVQLTRR740 pKa = 11.84KK741 pKa = 7.21TVTVEE746 pKa = 3.42KK747 pKa = 9.4TLNVQRR753 pKa = 11.84YY754 pKa = 5.43MVYY757 pKa = 9.35RR758 pKa = 11.84ALYY761 pKa = 9.0RR762 pKa = 11.84AYY764 pKa = 10.58ADD766 pKa = 3.4VTNNPMFGKK775 pKa = 10.58AMLTGFIFDD784 pKa = 4.31VLTQNAQLSAVAQLVSSAQNPMEE807 pKa = 4.14FLRR810 pKa = 11.84VISS813 pKa = 3.81
MM1 pKa = 7.34QEE3 pKa = 4.09RR4 pKa = 11.84DD5 pKa = 3.46SFTALDD11 pKa = 3.35ICDD14 pKa = 3.35GMRR17 pKa = 11.84EE18 pKa = 4.25GRR20 pKa = 11.84EE21 pKa = 3.51HH22 pKa = 5.93TFAVRR27 pKa = 11.84SGHH30 pKa = 5.73VPTGLRR36 pKa = 11.84ITCNGTYY43 pKa = 10.21LGVPFKK49 pKa = 10.56QAEE52 pKa = 4.21LLLIDD57 pKa = 4.57RR58 pKa = 11.84LPAKK62 pKa = 10.39LHH64 pKa = 5.7EE65 pKa = 5.19CNVAYY70 pKa = 10.06DD71 pKa = 3.63YY72 pKa = 11.65YY73 pKa = 9.94GTVVSARR80 pKa = 11.84LVPGRR85 pKa = 11.84DD86 pKa = 3.03ATYY89 pKa = 9.5VYY91 pKa = 10.72YY92 pKa = 10.88YY93 pKa = 10.38VDD95 pKa = 3.48QDD97 pKa = 3.76IQPVTQNLLGMLSRR111 pKa = 11.84HH112 pKa = 5.56FLGDD116 pKa = 3.14FSGYY120 pKa = 9.9YY121 pKa = 10.14NDD123 pKa = 3.8WCSLDD128 pKa = 3.39NVFYY132 pKa = 11.11SLSKK136 pKa = 10.45CDD138 pKa = 3.82NPVKK142 pKa = 10.39RR143 pKa = 11.84HH144 pKa = 5.12TIKK147 pKa = 10.22TIRR150 pKa = 11.84DD151 pKa = 3.42LPKK154 pKa = 9.78PKK156 pKa = 9.84ISGAHH161 pKa = 6.42HH162 pKa = 5.9IHH164 pKa = 5.41YY165 pKa = 7.07TASEE169 pKa = 4.26VWSVLDD175 pKa = 5.52DD176 pKa = 3.95RR177 pKa = 11.84GKK179 pKa = 10.59DD180 pKa = 3.43AARR183 pKa = 11.84HH184 pKa = 5.05ALRR187 pKa = 11.84LPADD191 pKa = 3.54ATTSFVGGVMLWLASLPTEE210 pKa = 4.15LLNPIIEE217 pKa = 4.53SDD219 pKa = 3.79LLDD222 pKa = 4.6AEE224 pKa = 4.45DD225 pKa = 4.19TIEE228 pKa = 4.36FGKK231 pKa = 10.36KK232 pKa = 9.3AKK234 pKa = 9.99KK235 pKa = 10.42LSVTAKK241 pKa = 9.18SFQNIVEE248 pKa = 4.23ADD250 pKa = 3.44LRR252 pKa = 11.84PIFEE256 pKa = 5.0ADD258 pKa = 3.32VLVNRR263 pKa = 11.84DD264 pKa = 3.22VGEE267 pKa = 4.06VDD269 pKa = 2.96WAGEE273 pKa = 4.0KK274 pKa = 10.67NNRR277 pKa = 11.84CKK279 pKa = 10.63PNLASVSPGEE289 pKa = 4.34VYY291 pKa = 10.91ARR293 pKa = 11.84TLALFTKK300 pKa = 10.34EE301 pKa = 4.44DD302 pKa = 3.95DD303 pKa = 3.92EE304 pKa = 4.6KK305 pKa = 11.4ALPRR309 pKa = 11.84RR310 pKa = 11.84LEE312 pKa = 3.92WEE314 pKa = 4.38KK315 pKa = 10.87FWDD318 pKa = 5.53ARR320 pKa = 11.84WQWSASGSIHH330 pKa = 6.31SQYY333 pKa = 10.91QEE335 pKa = 4.93DD336 pKa = 4.84LDD338 pKa = 4.13SLPKK342 pKa = 10.32EE343 pKa = 3.91RR344 pKa = 11.84EE345 pKa = 4.05LKK347 pKa = 10.82NKK349 pKa = 10.1FIALSMQGDD358 pKa = 3.82MPFEE362 pKa = 4.47HH363 pKa = 6.8FLNRR367 pKa = 11.84KK368 pKa = 8.12PEE370 pKa = 3.8IVAWSSVKK378 pKa = 10.74YY379 pKa = 9.63EE380 pKa = 3.89WGKK383 pKa = 9.08MRR385 pKa = 11.84AIYY388 pKa = 8.58GTDD391 pKa = 2.91LTSYY395 pKa = 10.79VLAHH399 pKa = 5.71YY400 pKa = 10.98AFFNCEE406 pKa = 3.81DD407 pKa = 4.08TLPTDD412 pKa = 4.19FPVGEE417 pKa = 4.21KK418 pKa = 10.28ARR420 pKa = 11.84PSFVSARR427 pKa = 11.84VAAILEE433 pKa = 4.51GTVPLCVDD441 pKa = 4.59FEE443 pKa = 4.98DD444 pKa = 5.9FNSQHH449 pKa = 6.83SNDD452 pKa = 3.2AMEE455 pKa = 5.1AVVQAYY461 pKa = 9.25IDD463 pKa = 4.37AYY465 pKa = 8.43SQYY468 pKa = 7.75MTPEE472 pKa = 4.02QVQAAMWTRR481 pKa = 11.84EE482 pKa = 4.39SISNTRR488 pKa = 11.84VIDD491 pKa = 3.62NMGTKK496 pKa = 9.69TSYY499 pKa = 8.55KK500 pKa = 9.92TKK502 pKa = 8.99GTLMSGWRR510 pKa = 11.84LTTFINSVLNYY521 pKa = 9.94VYY523 pKa = 9.67TQALISGSHH532 pKa = 5.12AMSRR536 pKa = 11.84SVHH539 pKa = 6.05NGDD542 pKa = 5.05DD543 pKa = 3.48VLLGIRR549 pKa = 11.84NFKK552 pKa = 9.78IVRR555 pKa = 11.84DD556 pKa = 3.79IVARR560 pKa = 11.84ADD562 pKa = 3.4KK563 pKa = 11.23YY564 pKa = 10.77NVRR567 pKa = 11.84LQRR570 pKa = 11.84TKK572 pKa = 10.81CAFGGLAEE580 pKa = 4.41FLRR583 pKa = 11.84VDD585 pKa = 4.33HH586 pKa = 6.34MRR588 pKa = 11.84GDD590 pKa = 3.57YY591 pKa = 10.53GQYY594 pKa = 8.72LTRR597 pKa = 11.84NIATVMHH604 pKa = 6.26SRR606 pKa = 11.84IEE608 pKa = 4.14SKK610 pKa = 10.49IAVSAVDD617 pKa = 3.43VVQAMEE623 pKa = 4.21DD624 pKa = 3.19RR625 pKa = 11.84LRR627 pKa = 11.84EE628 pKa = 3.81YY629 pKa = 10.48LQRR632 pKa = 11.84GGVWDD637 pKa = 3.61YY638 pKa = 11.72AVQLRR643 pKa = 11.84EE644 pKa = 3.98LYY646 pKa = 9.17YY647 pKa = 11.19DD648 pKa = 3.88RR649 pKa = 11.84MAPIYY654 pKa = 9.65SLRR657 pKa = 11.84PQDD660 pKa = 4.35LYY662 pKa = 11.27DD663 pKa = 3.14IRR665 pKa = 11.84TAHH668 pKa = 6.22RR669 pKa = 11.84VVGGIAEE676 pKa = 4.24DD677 pKa = 3.54VSARR681 pKa = 11.84VDD683 pKa = 3.23KK684 pKa = 10.52FIRR687 pKa = 11.84TSSEE691 pKa = 4.14SIEE694 pKa = 4.27TTLPDD699 pKa = 3.74TLPGVDD705 pKa = 3.73AYY707 pKa = 10.94ARR709 pKa = 11.84LLKK712 pKa = 10.36KK713 pKa = 10.55VLEE716 pKa = 4.35LEE718 pKa = 4.49VPVSVVSTRR727 pKa = 11.84IRR729 pKa = 11.84NATLNAVQLTRR740 pKa = 11.84KK741 pKa = 7.21TVTVEE746 pKa = 3.42KK747 pKa = 9.4TLNVQRR753 pKa = 11.84YY754 pKa = 5.43MVYY757 pKa = 9.35RR758 pKa = 11.84ALYY761 pKa = 9.0RR762 pKa = 11.84AYY764 pKa = 10.58ADD766 pKa = 3.4VTNNPMFGKK775 pKa = 10.58AMLTGFIFDD784 pKa = 4.31VLTQNAQLSAVAQLVSSAQNPMEE807 pKa = 4.14FLRR810 pKa = 11.84VISS813 pKa = 3.81
Molecular weight: 92.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1383 |
570 |
813 |
691.5 |
78.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.749 ± 0.19 | 1.085 ± 0.016 |
5.712 ± 0.854 | 6.146 ± 0.533 |
3.615 ± 0.054 | 5.423 ± 0.187 |
2.242 ± 0.02 | 5.061 ± 0.371 |
4.7 ± 0.019 | 8.894 ± 0.241 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.531 ± 0.038 | 3.543 ± 0.285 |
4.917 ± 0.891 | 3.398 ± 0.033 |
6.363 ± 0.471 | 7.158 ± 0.286 |
6.291 ± 0.102 | 7.737 ± 0.723 |
1.952 ± 0.257 | 4.483 ± 0.139 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
