
Sewage-associated circular DNA virus-14
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.03
Get precalculated fractions of proteins
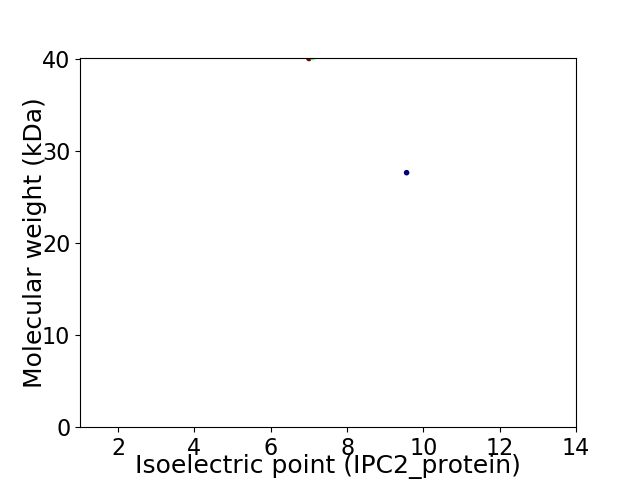
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075J175|A0A075J175_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-14 OX=1519390 PE=3 SV=1
MM1 pKa = 7.49EE2 pKa = 5.0VPPPAYY8 pKa = 10.0SPPPAAVSAEE18 pKa = 4.1VVPRR22 pKa = 11.84PFRR25 pKa = 11.84LAGKK29 pKa = 10.05AIFLTWPQNDD39 pKa = 3.32VSKK42 pKa = 10.66EE43 pKa = 3.71DD44 pKa = 3.61LMAKK48 pKa = 9.77IVALWEE54 pKa = 4.65AKK56 pKa = 10.51LSWAVVAEE64 pKa = 4.39EE65 pKa = 4.18SHH67 pKa = 7.01KK68 pKa = 11.02SGEE71 pKa = 4.23PHH73 pKa = 4.94VHH75 pKa = 7.05AIVQFAEE82 pKa = 4.2RR83 pKa = 11.84LDD85 pKa = 3.88LKK87 pKa = 10.69NANPVLDD94 pKa = 4.42ALTGKK99 pKa = 9.52HH100 pKa = 6.22GNYY103 pKa = 9.84QSVKK107 pKa = 9.21SAKK110 pKa = 9.23KK111 pKa = 9.2VLRR114 pKa = 11.84YY115 pKa = 9.18VCKK118 pKa = 9.43DD119 pKa = 3.09GQYY122 pKa = 8.29ITHH125 pKa = 6.79GEE127 pKa = 4.09VPDD130 pKa = 4.7FSEE133 pKa = 5.17KK134 pKa = 10.44PKK136 pKa = 10.32LQDD139 pKa = 2.75WAAKK143 pKa = 10.21LIIEE147 pKa = 4.2EE148 pKa = 4.1QKK150 pKa = 9.75TYY152 pKa = 10.82RR153 pKa = 11.84DD154 pKa = 3.65LVRR157 pKa = 11.84EE158 pKa = 3.98QPGFSMMQKK167 pKa = 9.99RR168 pKa = 11.84KK169 pKa = 10.15LEE171 pKa = 4.0EE172 pKa = 4.18FIGWNKK178 pKa = 9.05RR179 pKa = 11.84QKK181 pKa = 10.68LADD184 pKa = 3.68SLLPWRR190 pKa = 11.84VLTPKK195 pKa = 10.28PSASPVHH202 pKa = 5.21VALWTFLRR210 pKa = 11.84EE211 pKa = 3.99NVLVPRR217 pKa = 11.84TPRR220 pKa = 11.84QAQLWLCGLPGVGKK234 pKa = 8.44TRR236 pKa = 11.84FLAYY240 pKa = 10.08LRR242 pKa = 11.84ARR244 pKa = 11.84LRR246 pKa = 11.84VYY248 pKa = 11.12DD249 pKa = 3.88MPRR252 pKa = 11.84SEE254 pKa = 5.39DD255 pKa = 4.03FYY257 pKa = 11.69DD258 pKa = 4.1EE259 pKa = 5.17YY260 pKa = 11.45EE261 pKa = 4.63DD262 pKa = 4.01GCFDD266 pKa = 4.95LVVLDD271 pKa = 4.69EE272 pKa = 4.5YY273 pKa = 10.97KK274 pKa = 10.68AHH276 pKa = 7.45KK277 pKa = 9.94KK278 pKa = 9.65IQFLNAWADD287 pKa = 3.87GQPLPLRR294 pKa = 11.84QKK296 pKa = 10.86GSQLTKK302 pKa = 10.27TDD304 pKa = 3.96NLPLIIVSNYY314 pKa = 10.16SIEE317 pKa = 4.12EE318 pKa = 4.13VYY320 pKa = 10.66RR321 pKa = 11.84PGVGRR326 pKa = 11.84DD327 pKa = 3.3ALVDD331 pKa = 3.44RR332 pKa = 11.84FTQVFVDD339 pKa = 3.65TAFVIDD345 pKa = 4.41DD346 pKa = 4.26PEE348 pKa = 4.55HH349 pKa = 7.54VEE351 pKa = 3.9
MM1 pKa = 7.49EE2 pKa = 5.0VPPPAYY8 pKa = 10.0SPPPAAVSAEE18 pKa = 4.1VVPRR22 pKa = 11.84PFRR25 pKa = 11.84LAGKK29 pKa = 10.05AIFLTWPQNDD39 pKa = 3.32VSKK42 pKa = 10.66EE43 pKa = 3.71DD44 pKa = 3.61LMAKK48 pKa = 9.77IVALWEE54 pKa = 4.65AKK56 pKa = 10.51LSWAVVAEE64 pKa = 4.39EE65 pKa = 4.18SHH67 pKa = 7.01KK68 pKa = 11.02SGEE71 pKa = 4.23PHH73 pKa = 4.94VHH75 pKa = 7.05AIVQFAEE82 pKa = 4.2RR83 pKa = 11.84LDD85 pKa = 3.88LKK87 pKa = 10.69NANPVLDD94 pKa = 4.42ALTGKK99 pKa = 9.52HH100 pKa = 6.22GNYY103 pKa = 9.84QSVKK107 pKa = 9.21SAKK110 pKa = 9.23KK111 pKa = 9.2VLRR114 pKa = 11.84YY115 pKa = 9.18VCKK118 pKa = 9.43DD119 pKa = 3.09GQYY122 pKa = 8.29ITHH125 pKa = 6.79GEE127 pKa = 4.09VPDD130 pKa = 4.7FSEE133 pKa = 5.17KK134 pKa = 10.44PKK136 pKa = 10.32LQDD139 pKa = 2.75WAAKK143 pKa = 10.21LIIEE147 pKa = 4.2EE148 pKa = 4.1QKK150 pKa = 9.75TYY152 pKa = 10.82RR153 pKa = 11.84DD154 pKa = 3.65LVRR157 pKa = 11.84EE158 pKa = 3.98QPGFSMMQKK167 pKa = 9.99RR168 pKa = 11.84KK169 pKa = 10.15LEE171 pKa = 4.0EE172 pKa = 4.18FIGWNKK178 pKa = 9.05RR179 pKa = 11.84QKK181 pKa = 10.68LADD184 pKa = 3.68SLLPWRR190 pKa = 11.84VLTPKK195 pKa = 10.28PSASPVHH202 pKa = 5.21VALWTFLRR210 pKa = 11.84EE211 pKa = 3.99NVLVPRR217 pKa = 11.84TPRR220 pKa = 11.84QAQLWLCGLPGVGKK234 pKa = 8.44TRR236 pKa = 11.84FLAYY240 pKa = 10.08LRR242 pKa = 11.84ARR244 pKa = 11.84LRR246 pKa = 11.84VYY248 pKa = 11.12DD249 pKa = 3.88MPRR252 pKa = 11.84SEE254 pKa = 5.39DD255 pKa = 4.03FYY257 pKa = 11.69DD258 pKa = 4.1EE259 pKa = 5.17YY260 pKa = 11.45EE261 pKa = 4.63DD262 pKa = 4.01GCFDD266 pKa = 4.95LVVLDD271 pKa = 4.69EE272 pKa = 4.5YY273 pKa = 10.97KK274 pKa = 10.68AHH276 pKa = 7.45KK277 pKa = 9.94KK278 pKa = 9.65IQFLNAWADD287 pKa = 3.87GQPLPLRR294 pKa = 11.84QKK296 pKa = 10.86GSQLTKK302 pKa = 10.27TDD304 pKa = 3.96NLPLIIVSNYY314 pKa = 10.16SIEE317 pKa = 4.12EE318 pKa = 4.13VYY320 pKa = 10.66RR321 pKa = 11.84PGVGRR326 pKa = 11.84DD327 pKa = 3.3ALVDD331 pKa = 3.44RR332 pKa = 11.84FTQVFVDD339 pKa = 3.65TAFVIDD345 pKa = 4.41DD346 pKa = 4.26PEE348 pKa = 4.55HH349 pKa = 7.54VEE351 pKa = 3.9
Molecular weight: 40.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075J175|A0A075J175_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-14 OX=1519390 PE=3 SV=1
MM1 pKa = 7.38SMKK4 pKa = 10.56YY5 pKa = 9.96LGKK8 pKa = 10.4RR9 pKa = 11.84KK10 pKa = 7.95PTTWKK15 pKa = 9.77PRR17 pKa = 11.84SNPLTMMRR25 pKa = 11.84AKK27 pKa = 10.6ARR29 pKa = 11.84TSAFRR34 pKa = 11.84MTPRR38 pKa = 11.84APLSTRR44 pKa = 11.84GWRR47 pKa = 11.84PFISTNHH54 pKa = 5.24EE55 pKa = 4.23LKK57 pKa = 11.05VQDD60 pKa = 4.57LASATYY66 pKa = 9.84QVNTTGSITLLAVPITGADD85 pKa = 3.3YY86 pKa = 10.65NARR89 pKa = 11.84IGRR92 pKa = 11.84KK93 pKa = 7.46VTLKK97 pKa = 9.88SCFIRR102 pKa = 11.84GYY104 pKa = 11.01LRR106 pKa = 11.84IEE108 pKa = 4.23PGAATVGVAQAQQCRR123 pKa = 11.84FMLVLDD129 pKa = 4.53LQPNGAVFAITDD141 pKa = 4.51LLNTADD147 pKa = 4.18PTSQLNLNNRR157 pKa = 11.84DD158 pKa = 3.51RR159 pKa = 11.84FRR161 pKa = 11.84VLQDD165 pKa = 2.93QEE167 pKa = 3.85IAFDD171 pKa = 3.38HH172 pKa = 6.22FNYY175 pKa = 8.8VTTATQTSEE184 pKa = 3.86AFGRR188 pKa = 11.84TIQSFKK194 pKa = 10.52KK195 pKa = 10.21YY196 pKa = 10.17KK197 pKa = 10.23RR198 pKa = 11.84LNCEE202 pKa = 3.83MIFNAVNGGTIADD215 pKa = 3.76IASGALYY222 pKa = 8.82MVWIGSNAAGANTDD236 pKa = 3.42ANAVLSTRR244 pKa = 11.84VRR246 pKa = 11.84YY247 pKa = 9.79SDD249 pKa = 3.32SS250 pKa = 3.22
MM1 pKa = 7.38SMKK4 pKa = 10.56YY5 pKa = 9.96LGKK8 pKa = 10.4RR9 pKa = 11.84KK10 pKa = 7.95PTTWKK15 pKa = 9.77PRR17 pKa = 11.84SNPLTMMRR25 pKa = 11.84AKK27 pKa = 10.6ARR29 pKa = 11.84TSAFRR34 pKa = 11.84MTPRR38 pKa = 11.84APLSTRR44 pKa = 11.84GWRR47 pKa = 11.84PFISTNHH54 pKa = 5.24EE55 pKa = 4.23LKK57 pKa = 11.05VQDD60 pKa = 4.57LASATYY66 pKa = 9.84QVNTTGSITLLAVPITGADD85 pKa = 3.3YY86 pKa = 10.65NARR89 pKa = 11.84IGRR92 pKa = 11.84KK93 pKa = 7.46VTLKK97 pKa = 9.88SCFIRR102 pKa = 11.84GYY104 pKa = 11.01LRR106 pKa = 11.84IEE108 pKa = 4.23PGAATVGVAQAQQCRR123 pKa = 11.84FMLVLDD129 pKa = 4.53LQPNGAVFAITDD141 pKa = 4.51LLNTADD147 pKa = 4.18PTSQLNLNNRR157 pKa = 11.84DD158 pKa = 3.51RR159 pKa = 11.84FRR161 pKa = 11.84VLQDD165 pKa = 2.93QEE167 pKa = 3.85IAFDD171 pKa = 3.38HH172 pKa = 6.22FNYY175 pKa = 8.8VTTATQTSEE184 pKa = 3.86AFGRR188 pKa = 11.84TIQSFKK194 pKa = 10.52KK195 pKa = 10.21YY196 pKa = 10.17KK197 pKa = 10.23RR198 pKa = 11.84LNCEE202 pKa = 3.83MIFNAVNGGTIADD215 pKa = 3.76IASGALYY222 pKa = 8.82MVWIGSNAAGANTDD236 pKa = 3.42ANAVLSTRR244 pKa = 11.84VRR246 pKa = 11.84YY247 pKa = 9.79SDD249 pKa = 3.32SS250 pKa = 3.22
Molecular weight: 27.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
601 |
250 |
351 |
300.5 |
33.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.484 ± 1.046 | 0.998 ± 0.123 |
5.491 ± 0.665 | 4.825 ± 1.722 |
4.16 ± 0.146 | 5.158 ± 0.513 |
1.664 ± 0.527 | 4.16 ± 0.634 |
6.156 ± 1.071 | 9.318 ± 0.803 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.163 ± 0.632 | 4.16 ± 1.366 |
6.156 ± 1.315 | 4.493 ± 0.056 |
6.822 ± 0.718 | 5.491 ± 0.554 |
6.156 ± 2.343 | 7.82 ± 1.354 |
1.997 ± 0.486 | 3.328 ± 0.078 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
