
Thiohalophilus thiocyanatoxydans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Thioalkalispiraceae; Thiohalophilus
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
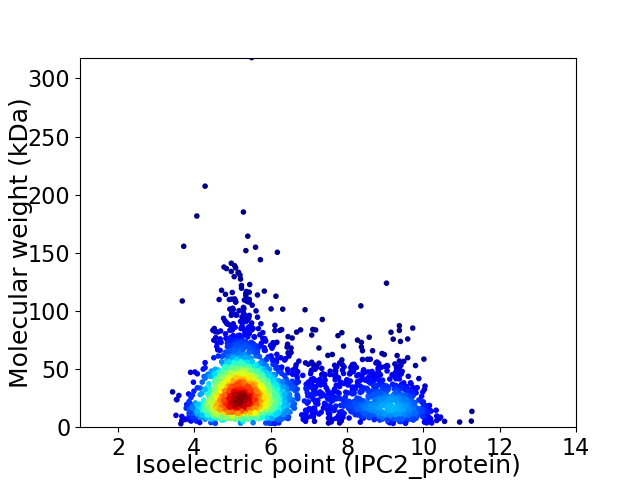
Virtual 2D-PAGE plot for 2775 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R8J2A3|A0A4R8J2A3_9GAMM LemA protein OS=Thiohalophilus thiocyanatoxydans OX=381308 GN=EDC23_0364 PE=3 SV=1
MM1 pKa = 7.52AGAMAQYY8 pKa = 10.08PDD10 pKa = 3.11ARR12 pKa = 11.84APFGLAGRR20 pKa = 11.84EE21 pKa = 3.89SRR23 pKa = 11.84LADD26 pKa = 3.31TLASLRR32 pKa = 11.84RR33 pKa = 11.84GWRR36 pKa = 11.84KK37 pKa = 5.96TARR40 pKa = 11.84IAGALLVGALLLAPAPSANAYY61 pKa = 8.69PADD64 pKa = 3.36QCAADD69 pKa = 4.57RR70 pKa = 11.84FGSDD74 pKa = 4.45LNCTANDD81 pKa = 3.52VAITNMQVIGDD92 pKa = 4.04MTWCEE97 pKa = 4.09GGTDD101 pKa = 2.96IVVDD105 pKa = 4.38LEE107 pKa = 4.49VTVNFASPDD116 pKa = 3.51RR117 pKa = 11.84NDD119 pKa = 2.92IGVFISNDD127 pKa = 3.19GNDD130 pKa = 3.89PQTTEE135 pKa = 4.19ANGGAASCSVSEE147 pKa = 5.25LPTEE151 pKa = 4.65SPFLDD156 pKa = 4.54LDD158 pKa = 4.06GDD160 pKa = 4.12SCGDD164 pKa = 3.57GNQSISGGTGSGTFYY179 pKa = 8.53MTDD182 pKa = 2.89VTVPCQAMPGSGNDD196 pKa = 3.32LYY198 pKa = 11.85VPFVVSWDD206 pKa = 3.49QDD208 pKa = 3.33KK209 pKa = 11.23QGQNVCSDD217 pKa = 3.19EE218 pKa = 4.31RR219 pKa = 11.84DD220 pKa = 3.43PVPRR224 pKa = 11.84TKK226 pKa = 10.45SKK228 pKa = 11.23CNAPDD233 pKa = 3.27IAQGTIQVGVMPSISKK249 pKa = 10.52SDD251 pKa = 3.96DD252 pKa = 2.94IEE254 pKa = 4.42YY255 pKa = 10.38IKK257 pKa = 11.21SGDD260 pKa = 3.48STTYY264 pKa = 11.12DD265 pKa = 3.23VTITNDD271 pKa = 3.1TGDD274 pKa = 3.83EE275 pKa = 4.03LGGAEE280 pKa = 4.33FSDD283 pKa = 4.04PAVSDD288 pKa = 3.54MSVDD292 pKa = 3.79SLNCTASGGATCPTGYY308 pKa = 9.23TIADD312 pKa = 3.67MQGDD316 pKa = 5.5GITLPDD322 pKa = 3.76MPDD325 pKa = 3.31GGSLTFEE332 pKa = 4.29IEE334 pKa = 3.88ATLTGDD340 pKa = 3.87PNDD343 pKa = 4.37TITNTGYY350 pKa = 8.79VTMGSHH356 pKa = 6.36TNSASDD362 pKa = 3.26TNTIVDD368 pKa = 4.88EE369 pKa = 4.48IGISPTSQNKK379 pKa = 9.12EE380 pKa = 3.51GAAGNTVTYY389 pKa = 9.75EE390 pKa = 3.83YY391 pKa = 10.05TVNNFTPWEE400 pKa = 4.18DD401 pKa = 4.66DD402 pKa = 3.01ISLSAVSSEE411 pKa = 3.53GWNVLLSDD419 pKa = 3.63ATLTVDD425 pKa = 4.52SEE427 pKa = 4.61GSADD431 pKa = 3.45FTLEE435 pKa = 3.78VEE437 pKa = 4.59IPAGASIDD445 pKa = 4.79DD446 pKa = 4.32FDD448 pKa = 4.24TTTITATSGDD458 pKa = 4.76DD459 pKa = 3.52PTMTASATAKK469 pKa = 7.12TTVAEE474 pKa = 4.37LLTLVPDD481 pKa = 3.58NTAAGGPGQTFSYY494 pKa = 10.61EE495 pKa = 3.72HH496 pKa = 6.37TLQNNFSSSQTISLSTAMTGDD517 pKa = 3.62CTDD520 pKa = 3.22WQATIYY526 pKa = 10.02EE527 pKa = 4.4ADD529 pKa = 3.78GEE531 pKa = 4.88TVISSVDD538 pKa = 3.44LDD540 pKa = 4.16GNGDD544 pKa = 3.41NTDD547 pKa = 2.88IVVEE551 pKa = 4.32VTAPNSAAIGDD562 pKa = 4.24SCVVTTTGTTSGVSASAKK580 pKa = 8.94DD581 pKa = 3.26TTTIKK586 pKa = 10.67EE587 pKa = 4.59LGLFWDD593 pKa = 4.02PDD595 pKa = 3.99YY596 pKa = 11.69EE597 pKa = 4.31EE598 pKa = 4.66EE599 pKa = 4.74SYY601 pKa = 10.52IYY603 pKa = 9.87PAGNNVYY610 pKa = 10.28GKK612 pKa = 10.71AFGLTDD618 pKa = 3.6GEE620 pKa = 4.42DD621 pKa = 4.84YY622 pKa = 11.14YY623 pKa = 11.42FEE625 pKa = 4.5WYY627 pKa = 10.07DD628 pKa = 3.38PAGDD632 pKa = 3.94LVRR635 pKa = 11.84TSSTTTVLGTVLPDD649 pKa = 3.47TYY651 pKa = 11.1KK652 pKa = 11.15LPDD655 pKa = 4.39DD656 pKa = 4.22ATLGTWNVKK665 pKa = 7.72VWHH668 pKa = 7.02DD669 pKa = 3.7TTPDD673 pKa = 3.5RR674 pKa = 11.84EE675 pKa = 4.33LHH677 pKa = 5.72QEE679 pKa = 3.43TDD681 pKa = 4.1FYY683 pKa = 11.56VGPDD687 pKa = 3.62HH688 pKa = 7.62IEE690 pKa = 4.03ASHH693 pKa = 7.26SGDD696 pKa = 3.96TEE698 pKa = 4.32TTDD701 pKa = 3.36TNVEE705 pKa = 3.73VDD707 pKa = 3.78LALHH711 pKa = 6.74DD712 pKa = 4.51RR713 pKa = 11.84FNHH716 pKa = 7.49AIPEE720 pKa = 4.25DD721 pKa = 4.08SNGDD725 pKa = 4.35LITANPPTTANPLLVTVTVDD745 pKa = 3.4GSASIVSTSLGNATISGQKK764 pKa = 8.13VTGLLDD770 pKa = 4.04DD771 pKa = 4.23TDD773 pKa = 4.13GTATMTITNSVAEE786 pKa = 4.46TVNVTPEE793 pKa = 3.79SHH795 pKa = 6.64EE796 pKa = 3.93PALYY800 pKa = 10.37GSPLRR805 pKa = 11.84DD806 pKa = 3.09EE807 pKa = 4.52SAAIDD812 pKa = 4.32FKK814 pKa = 11.21RR815 pKa = 11.84PPLDD819 pKa = 3.25HH820 pKa = 7.23FRR822 pKa = 11.84LEE824 pKa = 4.3YY825 pKa = 10.55NGQGLTCQPKK835 pKa = 9.92SITVKK840 pKa = 10.74ACIDD844 pKa = 3.76TNCNNLYY851 pKa = 10.43AYY853 pKa = 9.58DD854 pKa = 3.9VSVSLSPGGWVGGDD868 pKa = 3.2NKK870 pKa = 10.25VLSGGSEE877 pKa = 3.83EE878 pKa = 4.26FEE880 pKa = 4.31LRR882 pKa = 11.84KK883 pKa = 7.46TTTGTVTLDD892 pKa = 3.36IDD894 pKa = 4.14SSSPSPNNTPVCVNTATDD912 pKa = 4.11NEE914 pKa = 4.71SCDD917 pKa = 3.87LTFHH921 pKa = 7.8DD922 pKa = 5.28SGFLVDD928 pKa = 5.41VPTQTSCQTSGDD940 pKa = 3.77LTLSAVRR947 pKa = 11.84TDD949 pKa = 5.65DD950 pKa = 3.54VTQKK954 pKa = 10.34CVPAFASTTKK964 pKa = 10.53NLKK967 pKa = 9.18IWAEE971 pKa = 4.03YY972 pKa = 10.45SDD974 pKa = 4.44PASGTQSVEE983 pKa = 3.74LSHH986 pKa = 7.18AGNDD990 pKa = 3.41YY991 pKa = 10.95SLPDD995 pKa = 3.96TEE997 pKa = 5.4PGATNIPIEE1006 pKa = 4.29FNADD1010 pKa = 2.91AEE1012 pKa = 4.38ATYY1015 pKa = 11.46NLTYY1019 pKa = 10.14PDD1021 pKa = 3.69AGRR1024 pKa = 11.84LVLKK1028 pKa = 8.35TQYY1031 pKa = 10.25EE1032 pKa = 4.52GSGDD1036 pKa = 3.88DD1037 pKa = 4.97AGLTMLGDD1045 pKa = 3.68STYY1048 pKa = 8.98VTKK1051 pKa = 9.95PAKK1054 pKa = 10.34LYY1056 pKa = 10.01VHH1058 pKa = 6.9SDD1060 pKa = 3.36DD1061 pKa = 4.67TDD1063 pKa = 3.86ADD1065 pKa = 4.27CASGDD1070 pKa = 3.85ATCSRR1075 pKa = 11.84LTAAGDD1081 pKa = 3.74SFNMKK1086 pKa = 9.51VRR1088 pKa = 11.84GACADD1093 pKa = 3.4NSLTPNFRR1101 pKa = 11.84LDD1103 pKa = 5.3NIDD1106 pKa = 3.32LTQNLVAPAGGEE1118 pKa = 3.94SGSINVTNFNIADD1131 pKa = 3.86EE1132 pKa = 4.99DD1133 pKa = 4.0NGEE1136 pKa = 4.12HH1137 pKa = 7.32LINNQTVSEE1146 pKa = 4.31VGVFTFTASLGSGVDD1161 pKa = 3.67YY1162 pKa = 10.9FGEE1165 pKa = 4.18TSIGNTTLNTSDD1177 pKa = 3.48NIGRR1181 pKa = 11.84FSPHH1185 pKa = 6.2HH1186 pKa = 5.47FVVNATEE1193 pKa = 4.07ACNDD1197 pKa = 3.01SFTYY1201 pKa = 10.13SGQPFTVTAEE1211 pKa = 3.93ARR1213 pKa = 11.84NQGDD1217 pKa = 3.55NPTRR1221 pKa = 11.84NYY1223 pKa = 10.35RR1224 pKa = 11.84DD1225 pKa = 3.26GFVRR1229 pKa = 11.84DD1230 pKa = 3.88PVISNAGDD1238 pKa = 3.3TDD1240 pKa = 3.7NFSNNTLGSSEE1251 pKa = 4.01FTTNDD1256 pKa = 3.83GIGEE1260 pKa = 4.26RR1261 pKa = 11.84TDD1263 pKa = 2.9ITYY1266 pKa = 7.6TFPDD1270 pKa = 4.19KK1271 pKa = 10.17KK1272 pKa = 9.15TAPLNITEE1280 pKa = 4.66LRR1282 pKa = 11.84ATDD1285 pKa = 3.5SDD1287 pKa = 4.88GITSDD1292 pKa = 3.49TYY1294 pKa = 11.05TEE1296 pKa = 4.15GEE1298 pKa = 3.81INIRR1302 pKa = 11.84SGRR1305 pKa = 11.84IALDD1309 pKa = 3.19NAYY1312 pKa = 10.23GSEE1315 pKa = 4.06LVDD1318 pKa = 3.65LRR1320 pKa = 11.84VPMVAQYY1327 pKa = 11.32YY1328 pKa = 10.81DD1329 pKa = 3.01GDD1331 pKa = 3.73AFVRR1335 pKa = 11.84NEE1337 pKa = 4.69EE1338 pKa = 4.6DD1339 pKa = 3.05ICDD1342 pKa = 3.71NGVTISLLNPTDD1354 pKa = 3.58TLEE1357 pKa = 4.4VGDD1360 pKa = 4.21GEE1362 pKa = 4.82TRR1364 pKa = 11.84GEE1366 pKa = 4.28TCVQDD1371 pKa = 3.69TDD1373 pKa = 4.47DD1374 pKa = 5.02SGDD1377 pKa = 3.6SGAGCAIAADD1387 pKa = 3.99TDD1389 pKa = 3.54IQYY1392 pKa = 10.54TDD1394 pKa = 3.89APPDD1398 pKa = 3.61AGEE1401 pKa = 3.84YY1402 pKa = 9.45TLYY1405 pKa = 11.08LKK1407 pKa = 10.96APDD1410 pKa = 5.1DD1411 pKa = 4.95DD1412 pKa = 5.62SSFDD1416 pKa = 4.2PDD1418 pKa = 3.66DD1419 pKa = 5.16PIYY1422 pKa = 10.37GTLTVRR1428 pKa = 11.84ADD1430 pKa = 3.21APAYY1434 pKa = 9.78LQYY1437 pKa = 11.42DD1438 pKa = 3.54WDD1440 pKa = 4.64DD1441 pKa = 3.89DD1442 pKa = 3.92ATEE1445 pKa = 4.49EE1446 pKa = 4.34EE1447 pKa = 4.37PAATISFGRR1456 pKa = 11.84YY1457 pKa = 9.0RR1458 pKa = 11.84GDD1460 pKa = 3.36DD1461 pKa = 3.04RR1462 pKa = 11.84VIYY1465 pKa = 9.65WRR1467 pKa = 11.84EE1468 pKa = 3.59KK1469 pKa = 9.64FF1470 pKa = 3.49
MM1 pKa = 7.52AGAMAQYY8 pKa = 10.08PDD10 pKa = 3.11ARR12 pKa = 11.84APFGLAGRR20 pKa = 11.84EE21 pKa = 3.89SRR23 pKa = 11.84LADD26 pKa = 3.31TLASLRR32 pKa = 11.84RR33 pKa = 11.84GWRR36 pKa = 11.84KK37 pKa = 5.96TARR40 pKa = 11.84IAGALLVGALLLAPAPSANAYY61 pKa = 8.69PADD64 pKa = 3.36QCAADD69 pKa = 4.57RR70 pKa = 11.84FGSDD74 pKa = 4.45LNCTANDD81 pKa = 3.52VAITNMQVIGDD92 pKa = 4.04MTWCEE97 pKa = 4.09GGTDD101 pKa = 2.96IVVDD105 pKa = 4.38LEE107 pKa = 4.49VTVNFASPDD116 pKa = 3.51RR117 pKa = 11.84NDD119 pKa = 2.92IGVFISNDD127 pKa = 3.19GNDD130 pKa = 3.89PQTTEE135 pKa = 4.19ANGGAASCSVSEE147 pKa = 5.25LPTEE151 pKa = 4.65SPFLDD156 pKa = 4.54LDD158 pKa = 4.06GDD160 pKa = 4.12SCGDD164 pKa = 3.57GNQSISGGTGSGTFYY179 pKa = 8.53MTDD182 pKa = 2.89VTVPCQAMPGSGNDD196 pKa = 3.32LYY198 pKa = 11.85VPFVVSWDD206 pKa = 3.49QDD208 pKa = 3.33KK209 pKa = 11.23QGQNVCSDD217 pKa = 3.19EE218 pKa = 4.31RR219 pKa = 11.84DD220 pKa = 3.43PVPRR224 pKa = 11.84TKK226 pKa = 10.45SKK228 pKa = 11.23CNAPDD233 pKa = 3.27IAQGTIQVGVMPSISKK249 pKa = 10.52SDD251 pKa = 3.96DD252 pKa = 2.94IEE254 pKa = 4.42YY255 pKa = 10.38IKK257 pKa = 11.21SGDD260 pKa = 3.48STTYY264 pKa = 11.12DD265 pKa = 3.23VTITNDD271 pKa = 3.1TGDD274 pKa = 3.83EE275 pKa = 4.03LGGAEE280 pKa = 4.33FSDD283 pKa = 4.04PAVSDD288 pKa = 3.54MSVDD292 pKa = 3.79SLNCTASGGATCPTGYY308 pKa = 9.23TIADD312 pKa = 3.67MQGDD316 pKa = 5.5GITLPDD322 pKa = 3.76MPDD325 pKa = 3.31GGSLTFEE332 pKa = 4.29IEE334 pKa = 3.88ATLTGDD340 pKa = 3.87PNDD343 pKa = 4.37TITNTGYY350 pKa = 8.79VTMGSHH356 pKa = 6.36TNSASDD362 pKa = 3.26TNTIVDD368 pKa = 4.88EE369 pKa = 4.48IGISPTSQNKK379 pKa = 9.12EE380 pKa = 3.51GAAGNTVTYY389 pKa = 9.75EE390 pKa = 3.83YY391 pKa = 10.05TVNNFTPWEE400 pKa = 4.18DD401 pKa = 4.66DD402 pKa = 3.01ISLSAVSSEE411 pKa = 3.53GWNVLLSDD419 pKa = 3.63ATLTVDD425 pKa = 4.52SEE427 pKa = 4.61GSADD431 pKa = 3.45FTLEE435 pKa = 3.78VEE437 pKa = 4.59IPAGASIDD445 pKa = 4.79DD446 pKa = 4.32FDD448 pKa = 4.24TTTITATSGDD458 pKa = 4.76DD459 pKa = 3.52PTMTASATAKK469 pKa = 7.12TTVAEE474 pKa = 4.37LLTLVPDD481 pKa = 3.58NTAAGGPGQTFSYY494 pKa = 10.61EE495 pKa = 3.72HH496 pKa = 6.37TLQNNFSSSQTISLSTAMTGDD517 pKa = 3.62CTDD520 pKa = 3.22WQATIYY526 pKa = 10.02EE527 pKa = 4.4ADD529 pKa = 3.78GEE531 pKa = 4.88TVISSVDD538 pKa = 3.44LDD540 pKa = 4.16GNGDD544 pKa = 3.41NTDD547 pKa = 2.88IVVEE551 pKa = 4.32VTAPNSAAIGDD562 pKa = 4.24SCVVTTTGTTSGVSASAKK580 pKa = 8.94DD581 pKa = 3.26TTTIKK586 pKa = 10.67EE587 pKa = 4.59LGLFWDD593 pKa = 4.02PDD595 pKa = 3.99YY596 pKa = 11.69EE597 pKa = 4.31EE598 pKa = 4.66EE599 pKa = 4.74SYY601 pKa = 10.52IYY603 pKa = 9.87PAGNNVYY610 pKa = 10.28GKK612 pKa = 10.71AFGLTDD618 pKa = 3.6GEE620 pKa = 4.42DD621 pKa = 4.84YY622 pKa = 11.14YY623 pKa = 11.42FEE625 pKa = 4.5WYY627 pKa = 10.07DD628 pKa = 3.38PAGDD632 pKa = 3.94LVRR635 pKa = 11.84TSSTTTVLGTVLPDD649 pKa = 3.47TYY651 pKa = 11.1KK652 pKa = 11.15LPDD655 pKa = 4.39DD656 pKa = 4.22ATLGTWNVKK665 pKa = 7.72VWHH668 pKa = 7.02DD669 pKa = 3.7TTPDD673 pKa = 3.5RR674 pKa = 11.84EE675 pKa = 4.33LHH677 pKa = 5.72QEE679 pKa = 3.43TDD681 pKa = 4.1FYY683 pKa = 11.56VGPDD687 pKa = 3.62HH688 pKa = 7.62IEE690 pKa = 4.03ASHH693 pKa = 7.26SGDD696 pKa = 3.96TEE698 pKa = 4.32TTDD701 pKa = 3.36TNVEE705 pKa = 3.73VDD707 pKa = 3.78LALHH711 pKa = 6.74DD712 pKa = 4.51RR713 pKa = 11.84FNHH716 pKa = 7.49AIPEE720 pKa = 4.25DD721 pKa = 4.08SNGDD725 pKa = 4.35LITANPPTTANPLLVTVTVDD745 pKa = 3.4GSASIVSTSLGNATISGQKK764 pKa = 8.13VTGLLDD770 pKa = 4.04DD771 pKa = 4.23TDD773 pKa = 4.13GTATMTITNSVAEE786 pKa = 4.46TVNVTPEE793 pKa = 3.79SHH795 pKa = 6.64EE796 pKa = 3.93PALYY800 pKa = 10.37GSPLRR805 pKa = 11.84DD806 pKa = 3.09EE807 pKa = 4.52SAAIDD812 pKa = 4.32FKK814 pKa = 11.21RR815 pKa = 11.84PPLDD819 pKa = 3.25HH820 pKa = 7.23FRR822 pKa = 11.84LEE824 pKa = 4.3YY825 pKa = 10.55NGQGLTCQPKK835 pKa = 9.92SITVKK840 pKa = 10.74ACIDD844 pKa = 3.76TNCNNLYY851 pKa = 10.43AYY853 pKa = 9.58DD854 pKa = 3.9VSVSLSPGGWVGGDD868 pKa = 3.2NKK870 pKa = 10.25VLSGGSEE877 pKa = 3.83EE878 pKa = 4.26FEE880 pKa = 4.31LRR882 pKa = 11.84KK883 pKa = 7.46TTTGTVTLDD892 pKa = 3.36IDD894 pKa = 4.14SSSPSPNNTPVCVNTATDD912 pKa = 4.11NEE914 pKa = 4.71SCDD917 pKa = 3.87LTFHH921 pKa = 7.8DD922 pKa = 5.28SGFLVDD928 pKa = 5.41VPTQTSCQTSGDD940 pKa = 3.77LTLSAVRR947 pKa = 11.84TDD949 pKa = 5.65DD950 pKa = 3.54VTQKK954 pKa = 10.34CVPAFASTTKK964 pKa = 10.53NLKK967 pKa = 9.18IWAEE971 pKa = 4.03YY972 pKa = 10.45SDD974 pKa = 4.44PASGTQSVEE983 pKa = 3.74LSHH986 pKa = 7.18AGNDD990 pKa = 3.41YY991 pKa = 10.95SLPDD995 pKa = 3.96TEE997 pKa = 5.4PGATNIPIEE1006 pKa = 4.29FNADD1010 pKa = 2.91AEE1012 pKa = 4.38ATYY1015 pKa = 11.46NLTYY1019 pKa = 10.14PDD1021 pKa = 3.69AGRR1024 pKa = 11.84LVLKK1028 pKa = 8.35TQYY1031 pKa = 10.25EE1032 pKa = 4.52GSGDD1036 pKa = 3.88DD1037 pKa = 4.97AGLTMLGDD1045 pKa = 3.68STYY1048 pKa = 8.98VTKK1051 pKa = 9.95PAKK1054 pKa = 10.34LYY1056 pKa = 10.01VHH1058 pKa = 6.9SDD1060 pKa = 3.36DD1061 pKa = 4.67TDD1063 pKa = 3.86ADD1065 pKa = 4.27CASGDD1070 pKa = 3.85ATCSRR1075 pKa = 11.84LTAAGDD1081 pKa = 3.74SFNMKK1086 pKa = 9.51VRR1088 pKa = 11.84GACADD1093 pKa = 3.4NSLTPNFRR1101 pKa = 11.84LDD1103 pKa = 5.3NIDD1106 pKa = 3.32LTQNLVAPAGGEE1118 pKa = 3.94SGSINVTNFNIADD1131 pKa = 3.86EE1132 pKa = 4.99DD1133 pKa = 4.0NGEE1136 pKa = 4.12HH1137 pKa = 7.32LINNQTVSEE1146 pKa = 4.31VGVFTFTASLGSGVDD1161 pKa = 3.67YY1162 pKa = 10.9FGEE1165 pKa = 4.18TSIGNTTLNTSDD1177 pKa = 3.48NIGRR1181 pKa = 11.84FSPHH1185 pKa = 6.2HH1186 pKa = 5.47FVVNATEE1193 pKa = 4.07ACNDD1197 pKa = 3.01SFTYY1201 pKa = 10.13SGQPFTVTAEE1211 pKa = 3.93ARR1213 pKa = 11.84NQGDD1217 pKa = 3.55NPTRR1221 pKa = 11.84NYY1223 pKa = 10.35RR1224 pKa = 11.84DD1225 pKa = 3.26GFVRR1229 pKa = 11.84DD1230 pKa = 3.88PVISNAGDD1238 pKa = 3.3TDD1240 pKa = 3.7NFSNNTLGSSEE1251 pKa = 4.01FTTNDD1256 pKa = 3.83GIGEE1260 pKa = 4.26RR1261 pKa = 11.84TDD1263 pKa = 2.9ITYY1266 pKa = 7.6TFPDD1270 pKa = 4.19KK1271 pKa = 10.17KK1272 pKa = 9.15TAPLNITEE1280 pKa = 4.66LRR1282 pKa = 11.84ATDD1285 pKa = 3.5SDD1287 pKa = 4.88GITSDD1292 pKa = 3.49TYY1294 pKa = 11.05TEE1296 pKa = 4.15GEE1298 pKa = 3.81INIRR1302 pKa = 11.84SGRR1305 pKa = 11.84IALDD1309 pKa = 3.19NAYY1312 pKa = 10.23GSEE1315 pKa = 4.06LVDD1318 pKa = 3.65LRR1320 pKa = 11.84VPMVAQYY1327 pKa = 11.32YY1328 pKa = 10.81DD1329 pKa = 3.01GDD1331 pKa = 3.73AFVRR1335 pKa = 11.84NEE1337 pKa = 4.69EE1338 pKa = 4.6DD1339 pKa = 3.05ICDD1342 pKa = 3.71NGVTISLLNPTDD1354 pKa = 3.58TLEE1357 pKa = 4.4VGDD1360 pKa = 4.21GEE1362 pKa = 4.82TRR1364 pKa = 11.84GEE1366 pKa = 4.28TCVQDD1371 pKa = 3.69TDD1373 pKa = 4.47DD1374 pKa = 5.02SGDD1377 pKa = 3.6SGAGCAIAADD1387 pKa = 3.99TDD1389 pKa = 3.54IQYY1392 pKa = 10.54TDD1394 pKa = 3.89APPDD1398 pKa = 3.61AGEE1401 pKa = 3.84YY1402 pKa = 9.45TLYY1405 pKa = 11.08LKK1407 pKa = 10.96APDD1410 pKa = 5.1DD1411 pKa = 4.95DD1412 pKa = 5.62SSFDD1416 pKa = 4.2PDD1418 pKa = 3.66DD1419 pKa = 5.16PIYY1422 pKa = 10.37GTLTVRR1428 pKa = 11.84ADD1430 pKa = 3.21APAYY1434 pKa = 9.78LQYY1437 pKa = 11.42DD1438 pKa = 3.54WDD1440 pKa = 4.64DD1441 pKa = 3.89DD1442 pKa = 3.92ATEE1445 pKa = 4.49EE1446 pKa = 4.34EE1447 pKa = 4.37PAATISFGRR1456 pKa = 11.84YY1457 pKa = 9.0RR1458 pKa = 11.84GDD1460 pKa = 3.36DD1461 pKa = 3.04RR1462 pKa = 11.84VIYY1465 pKa = 9.65WRR1467 pKa = 11.84EE1468 pKa = 3.59KK1469 pKa = 9.64FF1470 pKa = 3.49
Molecular weight: 155.64 kDa
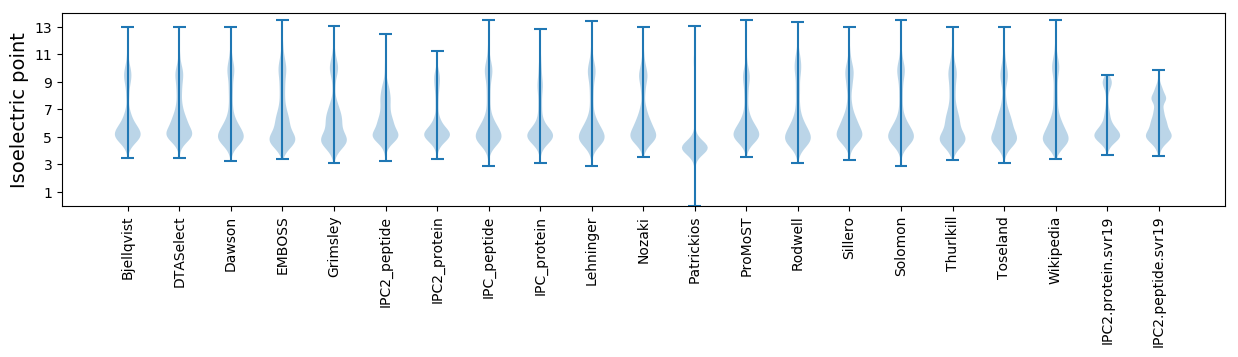
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R8J189|A0A4R8J189_9GAMM Cytochrome c oxidase cbb3-type subunit 2 OS=Thiohalophilus thiocyanatoxydans OX=381308 GN=EDC23_0310 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.99TRR25 pKa = 11.84GGRR28 pKa = 11.84KK29 pKa = 8.75VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.99TRR25 pKa = 11.84GGRR28 pKa = 11.84KK29 pKa = 8.75VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
832478 |
28 |
2858 |
300.0 |
33.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.419 ± 0.058 | 0.932 ± 0.017 |
5.899 ± 0.043 | 6.716 ± 0.056 |
3.555 ± 0.031 | 7.527 ± 0.051 |
2.533 ± 0.025 | 5.634 ± 0.037 |
3.658 ± 0.041 | 10.98 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.51 ± 0.024 | 3.234 ± 0.033 |
4.733 ± 0.031 | 4.557 ± 0.038 |
6.723 ± 0.045 | 5.351 ± 0.033 |
4.895 ± 0.033 | 6.859 ± 0.044 |
1.335 ± 0.02 | 2.95 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
