
Tistlia consotensis USBA 355
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhodovibrionaceae; Tistlia; Tistlia consotensis
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
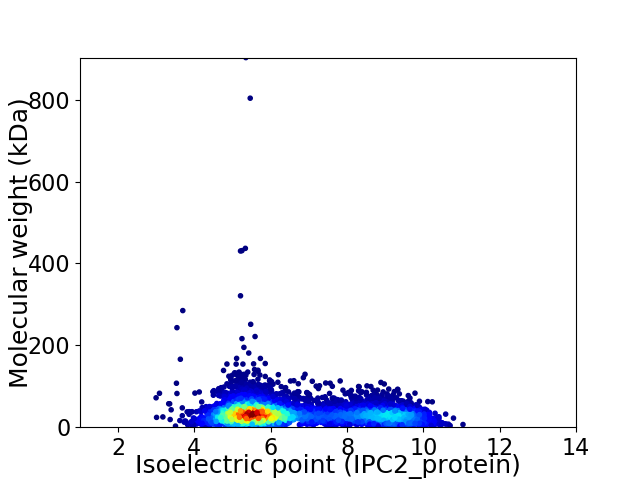
Virtual 2D-PAGE plot for 6388 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y6BC91|A0A1Y6BC91_9PROT Acetolactate synthase large subunit OS=Tistlia consotensis USBA 355 OX=560819 GN=SAMN05428998_103128 PE=3 SV=1
EEE2 pKa = 5.37HH3 pKa = 6.97YY4 pKa = 10.97YY5 pKa = 10.8DD6 pKa = 5.11DD7 pKa = 5.21SGATDDD13 pKa = 4.53LQNSQNLLFDDD24 pKa = 4.67IGANGYYY31 pKa = 9.94DD32 pKa = 3.28NAYYY36 pKa = 10.25NYYY39 pKa = 9.48DD40 pKa = 3.23SLTVTDDD47 pKa = 3.99KK48 pKa = 11.64SGAVVGEEE56 pKa = 3.67DDD58 pKa = 3.44LVRR61 pKa = 11.84VAPDDD66 pKa = 3.01VVAADDD72 pKa = 3.6SGDDD76 pKa = 3.58YY77 pKa = 7.91TIQAAIDDD85 pKa = 3.44AQAGDDD91 pKa = 4.25IVVAAGSYYY100 pKa = 9.56PGLVDDD106 pKa = 4.6YY107 pKa = 9.06TASFGAGRR115 pKa = 11.84GTLNQQILVDDD126 pKa = 4.35KK127 pKa = 10.48LTLIGLPGATIQVAEEE143 pKa = 4.48DDD145 pKa = 3.74TALADDD151 pKa = 3.79TLGFTVAADDD161 pKa = 3.76DD162 pKa = 3.93SISGFEEE169 pKa = 3.84VGPLGDDD176 pKa = 3.79QQDDD180 pKa = 3.31FDDD183 pKa = 3.68ANRR186 pKa = 11.84GYYY189 pKa = 11.03YYY191 pKa = 10.62IFVDDD196 pKa = 4.23EE197 pKa = 4.75VLGTTLSGNTIQDDD211 pKa = 3.48RR212 pKa = 11.84TGITFEEE219 pKa = 4.23ANGDDD224 pKa = 3.99LDDD227 pKa = 4.4SHHH230 pKa = 6.38TSVTGNEEE238 pKa = 3.79DDD240 pKa = 3.54TRR242 pKa = 11.84GAFLVRR248 pKa = 11.84SEEE251 pKa = 4.12VEEE254 pKa = 3.81SDDD257 pKa = 4.35SFGAEEE263 pKa = 3.78NEEE266 pKa = 3.56DDD268 pKa = 4.07TILAGTQPGDDD279 pKa = 3.64YY280 pKa = 10.31ADDD283 pKa = 3.89LVDDD287 pKa = 3.41AQYYY291 pKa = 10.91DDD293 pKa = 4.55DD294 pKa = 3.65MALSTANDDD303 pKa = 2.91MTIVDDD309 pKa = 3.35RR310 pKa = 11.84YYY312 pKa = 10.76EEE314 pKa = 4.55GVLTQQYYY322 pKa = 10.63SSDDD326 pKa = 3.68ALSQQYYY333 pKa = 10.34AIANRR338 pKa = 11.84THHH341 pKa = 7.17EEE343 pKa = 3.92RR344 pKa = 11.84AGADDD349 pKa = 3.61DD350 pKa = 4.0PSAALGEEE358 pKa = 4.34RR359 pKa = 11.84GNGFGSEEE367 pKa = 4.02LPIGSLQDDD376 pKa = 3.35VDDD379 pKa = 3.52VVKKK383 pKa = 10.23GSVHHH388 pKa = 6.48QAGDDD393 pKa = 3.68YY394 pKa = 10.88GEEE397 pKa = 4.24VAVATEEE404 pKa = 3.92LTVDDD409 pKa = 4.13DDD411 pKa = 3.68GAIGIALQLASGIEEE426 pKa = 4.1VTATGAADDD435 pKa = 3.39ALTGNEEE442 pKa = 4.18ANLLAGGEEE451 pKa = 4.2DDD453 pKa = 4.52DD454 pKa = 3.72LFGGGAADDD463 pKa = 3.98LTGGGGNDDD472 pKa = 3.31LVGGEEE478 pKa = 4.5TDDD481 pKa = 3.4AHHH484 pKa = 6.15TGAQADDD491 pKa = 3.95YY492 pKa = 11.02IALNDDD498 pKa = 4.12DD499 pKa = 3.82SFTVADDD506 pKa = 5.02SAGRR510 pKa = 11.84DDD512 pKa = 3.72SDDD515 pKa = 2.91HH516 pKa = 6.8SGVEEE521 pKa = 3.67LAFGDDD527 pKa = 4.21QVLLVGGSGFDDD539 pKa = 3.59LQGAIDDD546 pKa = 3.8ASDDD550 pKa = 3.44DDD552 pKa = 4.29ILVYYY557 pKa = 10.24GQYYY561 pKa = 9.81EEE563 pKa = 4.34ANYYY567 pKa = 9.71PNTGLNDDD575 pKa = 3.57SYYY578 pKa = 11.49GGNNPLGLVIGKKK591 pKa = 9.29VTIQGVDDD599 pKa = 3.4AGHHH603 pKa = 7.59ITDDD607 pKa = 3.64HH608 pKa = 6.61GVEEE612 pKa = 4.22TISATRR618 pKa = 11.84QSNFGTSFLVTADDD632 pKa = 3.22HH633 pKa = 6.26AIHHH637 pKa = 5.88LEEE640 pKa = 3.94LGNSSPNQPYYY651 pKa = 8.28NKKK654 pKa = 9.64IEEE657 pKa = 4.43IADDD661 pKa = 4.1FTLDDD666 pKa = 3.57SVVGAVDDD674 pKa = 3.52YY675 pKa = 11.49HHH677 pKa = 7.38HHH679 pKa = 4.92QPTAADDD686 pKa = 3.8LSGSWDDD693 pKa = 3.31HHH695 pKa = 4.18QMLTAVYYY703 pKa = 10.22NDDD706 pKa = 3.67YYY708 pKa = 10.53TVPSDDD714 pKa = 4.23AGWQSAISSYYY725 pKa = 11.09DD726 pKa = 3.35EEE728 pKa = 4.82SQLAGTIEEE737 pKa = 4.33ASGPGYYY744 pKa = 10.79YYY746 pKa = 10.99ADDD749 pKa = 3.86LDDD752 pKa = 3.54RR753 pKa = 11.84IVGNLFINVAADDD766 pKa = 3.39DD767 pKa = 4.34TWHHH771 pKa = 6.68NGILITGQDDD781 pKa = 3.63EEE783 pKa = 4.65GWRR786 pKa = 11.84LAPAALPTEEE796 pKa = 3.71SGNVFGADDD805 pKa = 2.99DDD807 pKa = 4.51IFWVRR812 pKa = 11.84GEEE815 pKa = 4.31AQDDD819 pKa = 3.96PDDD822 pKa = 3.7AYYY825 pKa = 10.49QSLIANNDDD834 pKa = 3.3PAYYY838 pKa = 9.6YYY840 pKa = 10.34VDDD843 pKa = 4.03SGAPAIGTYYY853 pKa = 10.63DD854 pKa = 3.63DDD856 pKa = 4.5GPLLPGGVPVTPSSPSFAIRR876 pKa = 11.84QSAGDDD882 pKa = 3.43DDD884 pKa = 6.2NEEE887 pKa = 3.88HHH889 pKa = 6.93ADDD892 pKa = 3.4LVVKKK897 pKa = 10.63EEE899 pKa = 3.69DDD901 pKa = 3.15ASRR904 pKa = 11.84TLSLIVGSDDD914 pKa = 3.3ADDD917 pKa = 3.27LSGGSGDDD925 pKa = 3.66EE926 pKa = 4.02LLGGGGDDD934 pKa = 3.97DD935 pKa = 5.2IDDD938 pKa = 3.67GGGNDDD944 pKa = 5.25ILGGAGDDD952 pKa = 4.33DD953 pKa = 3.87LFGGAGNDDD962 pKa = 3.55LAGGDDD968 pKa = 3.61SDDD971 pKa = 3.74VTGGQGDDD979 pKa = 4.18DD980 pKa = 3.04VVFQQGEEE988 pKa = 4.34GSDDD992 pKa = 3.12VTDDD996 pKa = 3.6SGGDDD1001 pKa = 3.68LDDD1004 pKa = 4.18SDDD1007 pKa = 5.19VQSDDD1012 pKa = 4.36PAEEE1016 pKa = 3.92HHH1018 pKa = 7.02GDDD1021 pKa = 4.01TDDD1024 pKa = 2.81HHH1026 pKa = 5.57QVATAAAPEEE1036 pKa = 4.28PLAVVEEE1043 pKa = 4.42VPAASLTVDDD1053 pKa = 2.8HH1054 pKa = 7.57DD1055 pKa = 4.88NVTLTAAA
EEE2 pKa = 5.37HH3 pKa = 6.97YY4 pKa = 10.97YY5 pKa = 10.8DD6 pKa = 5.11DD7 pKa = 5.21SGATDDD13 pKa = 4.53LQNSQNLLFDDD24 pKa = 4.67IGANGYYY31 pKa = 9.94DD32 pKa = 3.28NAYYY36 pKa = 10.25NYYY39 pKa = 9.48DD40 pKa = 3.23SLTVTDDD47 pKa = 3.99KK48 pKa = 11.64SGAVVGEEE56 pKa = 3.67DDD58 pKa = 3.44LVRR61 pKa = 11.84VAPDDD66 pKa = 3.01VVAADDD72 pKa = 3.6SGDDD76 pKa = 3.58YY77 pKa = 7.91TIQAAIDDD85 pKa = 3.44AQAGDDD91 pKa = 4.25IVVAAGSYYY100 pKa = 9.56PGLVDDD106 pKa = 4.6YY107 pKa = 9.06TASFGAGRR115 pKa = 11.84GTLNQQILVDDD126 pKa = 4.35KK127 pKa = 10.48LTLIGLPGATIQVAEEE143 pKa = 4.48DDD145 pKa = 3.74TALADDD151 pKa = 3.79TLGFTVAADDD161 pKa = 3.76DD162 pKa = 3.93SISGFEEE169 pKa = 3.84VGPLGDDD176 pKa = 3.79QQDDD180 pKa = 3.31FDDD183 pKa = 3.68ANRR186 pKa = 11.84GYYY189 pKa = 11.03YYY191 pKa = 10.62IFVDDD196 pKa = 4.23EE197 pKa = 4.75VLGTTLSGNTIQDDD211 pKa = 3.48RR212 pKa = 11.84TGITFEEE219 pKa = 4.23ANGDDD224 pKa = 3.99LDDD227 pKa = 4.4SHHH230 pKa = 6.38TSVTGNEEE238 pKa = 3.79DDD240 pKa = 3.54TRR242 pKa = 11.84GAFLVRR248 pKa = 11.84SEEE251 pKa = 4.12VEEE254 pKa = 3.81SDDD257 pKa = 4.35SFGAEEE263 pKa = 3.78NEEE266 pKa = 3.56DDD268 pKa = 4.07TILAGTQPGDDD279 pKa = 3.64YY280 pKa = 10.31ADDD283 pKa = 3.89LVDDD287 pKa = 3.41AQYYY291 pKa = 10.91DDD293 pKa = 4.55DD294 pKa = 3.65MALSTANDDD303 pKa = 2.91MTIVDDD309 pKa = 3.35RR310 pKa = 11.84YYY312 pKa = 10.76EEE314 pKa = 4.55GVLTQQYYY322 pKa = 10.63SSDDD326 pKa = 3.68ALSQQYYY333 pKa = 10.34AIANRR338 pKa = 11.84THHH341 pKa = 7.17EEE343 pKa = 3.92RR344 pKa = 11.84AGADDD349 pKa = 3.61DD350 pKa = 4.0PSAALGEEE358 pKa = 4.34RR359 pKa = 11.84GNGFGSEEE367 pKa = 4.02LPIGSLQDDD376 pKa = 3.35VDDD379 pKa = 3.52VVKKK383 pKa = 10.23GSVHHH388 pKa = 6.48QAGDDD393 pKa = 3.68YY394 pKa = 10.88GEEE397 pKa = 4.24VAVATEEE404 pKa = 3.92LTVDDD409 pKa = 4.13DDD411 pKa = 3.68GAIGIALQLASGIEEE426 pKa = 4.1VTATGAADDD435 pKa = 3.39ALTGNEEE442 pKa = 4.18ANLLAGGEEE451 pKa = 4.2DDD453 pKa = 4.52DD454 pKa = 3.72LFGGGAADDD463 pKa = 3.98LTGGGGNDDD472 pKa = 3.31LVGGEEE478 pKa = 4.5TDDD481 pKa = 3.4AHHH484 pKa = 6.15TGAQADDD491 pKa = 3.95YY492 pKa = 11.02IALNDDD498 pKa = 4.12DD499 pKa = 3.82SFTVADDD506 pKa = 5.02SAGRR510 pKa = 11.84DDD512 pKa = 3.72SDDD515 pKa = 2.91HH516 pKa = 6.8SGVEEE521 pKa = 3.67LAFGDDD527 pKa = 4.21QVLLVGGSGFDDD539 pKa = 3.59LQGAIDDD546 pKa = 3.8ASDDD550 pKa = 3.44DDD552 pKa = 4.29ILVYYY557 pKa = 10.24GQYYY561 pKa = 9.81EEE563 pKa = 4.34ANYYY567 pKa = 9.71PNTGLNDDD575 pKa = 3.57SYYY578 pKa = 11.49GGNNPLGLVIGKKK591 pKa = 9.29VTIQGVDDD599 pKa = 3.4AGHHH603 pKa = 7.59ITDDD607 pKa = 3.64HH608 pKa = 6.61GVEEE612 pKa = 4.22TISATRR618 pKa = 11.84QSNFGTSFLVTADDD632 pKa = 3.22HH633 pKa = 6.26AIHHH637 pKa = 5.88LEEE640 pKa = 3.94LGNSSPNQPYYY651 pKa = 8.28NKKK654 pKa = 9.64IEEE657 pKa = 4.43IADDD661 pKa = 4.1FTLDDD666 pKa = 3.57SVVGAVDDD674 pKa = 3.52YY675 pKa = 11.49HHH677 pKa = 7.38HHH679 pKa = 4.92QPTAADDD686 pKa = 3.8LSGSWDDD693 pKa = 3.31HHH695 pKa = 4.18QMLTAVYYY703 pKa = 10.22NDDD706 pKa = 3.67YYY708 pKa = 10.53TVPSDDD714 pKa = 4.23AGWQSAISSYYY725 pKa = 11.09DD726 pKa = 3.35EEE728 pKa = 4.82SQLAGTIEEE737 pKa = 4.33ASGPGYYY744 pKa = 10.79YYY746 pKa = 10.99ADDD749 pKa = 3.86LDDD752 pKa = 3.54RR753 pKa = 11.84IVGNLFINVAADDD766 pKa = 3.39DD767 pKa = 4.34TWHHH771 pKa = 6.68NGILITGQDDD781 pKa = 3.63EEE783 pKa = 4.65GWRR786 pKa = 11.84LAPAALPTEEE796 pKa = 3.71SGNVFGADDD805 pKa = 2.99DDD807 pKa = 4.51IFWVRR812 pKa = 11.84GEEE815 pKa = 4.31AQDDD819 pKa = 3.96PDDD822 pKa = 3.7AYYY825 pKa = 10.49QSLIANNDDD834 pKa = 3.3PAYYY838 pKa = 9.6YYY840 pKa = 10.34VDDD843 pKa = 4.03SGAPAIGTYYY853 pKa = 10.63DD854 pKa = 3.63DDD856 pKa = 4.5GPLLPGGVPVTPSSPSFAIRR876 pKa = 11.84QSAGDDD882 pKa = 3.43DDD884 pKa = 6.2NEEE887 pKa = 3.88HHH889 pKa = 6.93ADDD892 pKa = 3.4LVVKKK897 pKa = 10.63EEE899 pKa = 3.69DDD901 pKa = 3.15ASRR904 pKa = 11.84TLSLIVGSDDD914 pKa = 3.3ADDD917 pKa = 3.27LSGGSGDDD925 pKa = 3.66EE926 pKa = 4.02LLGGGGDDD934 pKa = 3.97DD935 pKa = 5.2IDDD938 pKa = 3.67GGGNDDD944 pKa = 5.25ILGGAGDDD952 pKa = 4.33DD953 pKa = 3.87LFGGAGNDDD962 pKa = 3.55LAGGDDD968 pKa = 3.61SDDD971 pKa = 3.74VTGGQGDDD979 pKa = 4.18DD980 pKa = 3.04VVFQQGEEE988 pKa = 4.34GSDDD992 pKa = 3.12VTDDD996 pKa = 3.6SGGDDD1001 pKa = 3.68LDDD1004 pKa = 4.18SDDD1007 pKa = 5.19VQSDDD1012 pKa = 4.36PAEEE1016 pKa = 3.92HHH1018 pKa = 7.02GDDD1021 pKa = 4.01TDDD1024 pKa = 2.81HHH1026 pKa = 5.57QVATAAAPEEE1036 pKa = 4.28PLAVVEEE1043 pKa = 4.42VPAASLTVDDD1053 pKa = 2.8HH1054 pKa = 7.57DD1055 pKa = 4.88NVTLTAAA
Molecular weight: 107.38 kDa
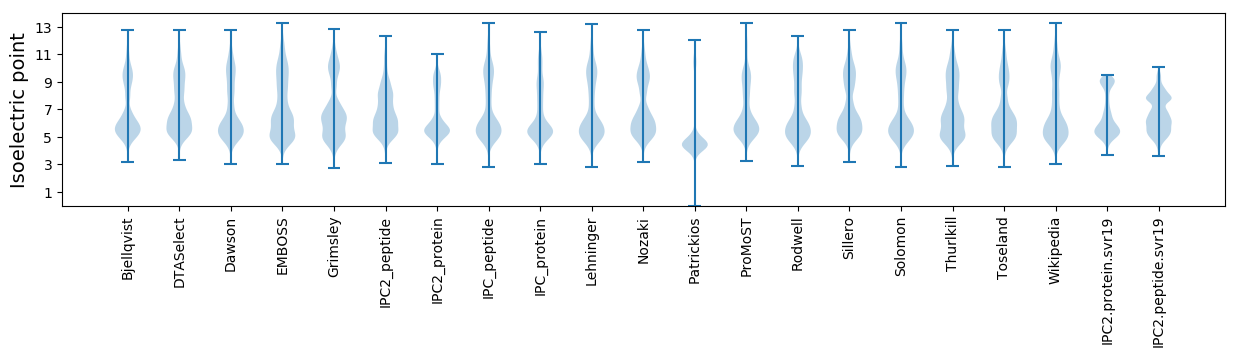
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y6CEB2|A0A1Y6CEB2_9PROT Crotonobetaine/carnitine-CoA ligase OS=Tistlia consotensis USBA 355 OX=560819 GN=SAMN05428998_12153 PE=4 SV=1
MM1 pKa = 7.59RR2 pKa = 11.84RR3 pKa = 11.84LRR5 pKa = 11.84RR6 pKa = 11.84LLGLAVAGQPVPRR19 pKa = 11.84THH21 pKa = 7.39GLGLLRR27 pKa = 11.84AAWAGRR33 pKa = 11.84GRR35 pKa = 11.84LLRR38 pKa = 11.84ALAGTIRR45 pKa = 11.84PVRR48 pKa = 11.84RR49 pKa = 11.84GRR51 pKa = 11.84TRR53 pKa = 11.84EE54 pKa = 3.83PRR56 pKa = 11.84PVAGG60 pKa = 4.3
MM1 pKa = 7.59RR2 pKa = 11.84RR3 pKa = 11.84LRR5 pKa = 11.84RR6 pKa = 11.84LLGLAVAGQPVPRR19 pKa = 11.84THH21 pKa = 7.39GLGLLRR27 pKa = 11.84AAWAGRR33 pKa = 11.84GRR35 pKa = 11.84LLRR38 pKa = 11.84ALAGTIRR45 pKa = 11.84PVRR48 pKa = 11.84RR49 pKa = 11.84GRR51 pKa = 11.84TRR53 pKa = 11.84EE54 pKa = 3.83PRR56 pKa = 11.84PVAGG60 pKa = 4.3
Molecular weight: 6.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2078936 |
25 |
8553 |
325.4 |
35.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.882 ± 0.049 | 0.896 ± 0.011 |
5.291 ± 0.023 | 6.143 ± 0.035 |
3.474 ± 0.02 | 9.292 ± 0.029 |
1.857 ± 0.014 | 4.004 ± 0.019 |
2.426 ± 0.025 | 11.638 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.952 ± 0.016 | 1.899 ± 0.016 |
5.697 ± 0.026 | 2.955 ± 0.017 |
8.126 ± 0.037 | 4.856 ± 0.026 |
4.634 ± 0.03 | 7.426 ± 0.026 |
1.424 ± 0.012 | 2.127 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
