
Beet curly top Iran virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Becurtovirus
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
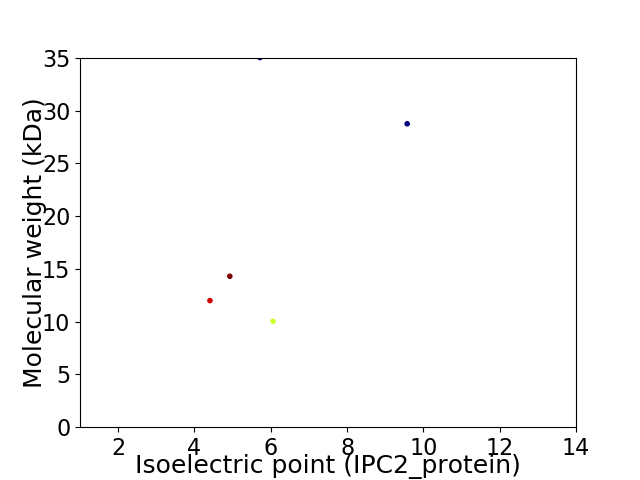
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
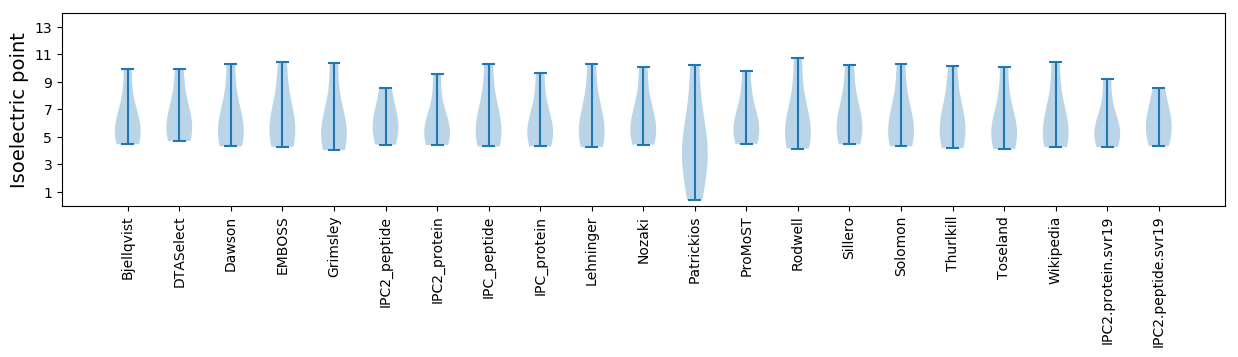
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L7XB51|L7XB51_9GEMI Ss-ds DNA regulator OS=Beet curly top Iran virus OX=391228 GN=V3 PE=4 SV=1
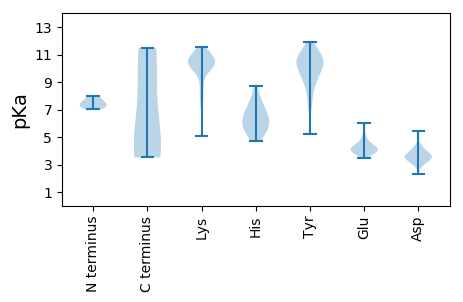
MM1 pKa = 8.0SGMVLDD7 pKa = 4.27WSLYY11 pKa = 10.53DD12 pKa = 3.81IDD14 pKa = 3.83HH15 pKa = 5.88TTYY18 pKa = 10.67HH19 pKa = 6.85IIDD22 pKa = 5.02DD23 pKa = 3.38IRR25 pKa = 11.84YY26 pKa = 9.29QKK28 pKa = 9.96IQQEE32 pKa = 4.35LFKK35 pKa = 11.14SIIGCNEE42 pKa = 4.3DD43 pKa = 3.19YY44 pKa = 11.11SVWIKK49 pKa = 10.7HH50 pKa = 5.77KK51 pKa = 9.99PNLVIPGGRR60 pKa = 11.84PCIAITNPDD69 pKa = 4.09MDD71 pKa = 5.47WIPCMSEE78 pKa = 3.98SMKK81 pKa = 10.99DD82 pKa = 2.94WFYY85 pKa = 11.88ANCDD89 pKa = 3.2VYY91 pKa = 11.63YY92 pKa = 10.6LANDD96 pKa = 3.86EE97 pKa = 4.12VWYY100 pKa = 8.89TT101 pKa = 3.52
MM1 pKa = 8.0SGMVLDD7 pKa = 4.27WSLYY11 pKa = 10.53DD12 pKa = 3.81IDD14 pKa = 3.83HH15 pKa = 5.88TTYY18 pKa = 10.67HH19 pKa = 6.85IIDD22 pKa = 5.02DD23 pKa = 3.38IRR25 pKa = 11.84YY26 pKa = 9.29QKK28 pKa = 9.96IQQEE32 pKa = 4.35LFKK35 pKa = 11.14SIIGCNEE42 pKa = 4.3DD43 pKa = 3.19YY44 pKa = 11.11SVWIKK49 pKa = 10.7HH50 pKa = 5.77KK51 pKa = 9.99PNLVIPGGRR60 pKa = 11.84PCIAITNPDD69 pKa = 4.09MDD71 pKa = 5.47WIPCMSEE78 pKa = 3.98SMKK81 pKa = 10.99DD82 pKa = 2.94WFYY85 pKa = 11.88ANCDD89 pKa = 3.2VYY91 pKa = 11.63YY92 pKa = 10.6LANDD96 pKa = 3.86EE97 pKa = 4.12VWYY100 pKa = 8.89TT101 pKa = 3.52
Molecular weight: 11.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L7XDF9|L7XDF9_9GEMI Capsid protein OS=Beet curly top Iran virus OX=391228 GN=V1 PE=3 SV=1
MM1 pKa = 7.59AVQSQKK7 pKa = 10.4RR8 pKa = 11.84KK9 pKa = 10.22YY10 pKa = 9.69MSATNWAKK18 pKa = 10.31KK19 pKa = 10.17RR20 pKa = 11.84KK21 pKa = 5.06TTTGRR26 pKa = 11.84TVSKK30 pKa = 10.05KK31 pKa = 8.12YY32 pKa = 7.44QWKK35 pKa = 10.13RR36 pKa = 11.84PVRR39 pKa = 11.84SNRR42 pKa = 11.84SVKK45 pKa = 10.6LKK47 pKa = 10.23MYY49 pKa = 10.74DD50 pKa = 3.68DD51 pKa = 4.07MLGASGVGSTISNDD65 pKa = 3.59GMITMLNNYY74 pKa = 7.83VQGIGDD80 pKa = 3.83SQRR83 pKa = 11.84SNNLTVTRR91 pKa = 11.84HH92 pKa = 5.78LKK94 pKa = 10.43FDD96 pKa = 3.68MALMASSMFWEE107 pKa = 4.88RR108 pKa = 11.84PNYY111 pKa = 5.2MTQYY115 pKa = 9.32HH116 pKa = 5.44WVIVDD121 pKa = 3.71RR122 pKa = 11.84DD123 pKa = 3.63VGATFPTQLSTIFDD137 pKa = 3.94IPTGGQAMPSTYY149 pKa = 10.12RR150 pKa = 11.84IRR152 pKa = 11.84RR153 pKa = 11.84DD154 pKa = 3.09ANEE157 pKa = 3.67RR158 pKa = 11.84FIVKK162 pKa = 10.08KK163 pKa = 10.17KK164 pKa = 8.49WQTHH168 pKa = 4.72LMSSGTDD175 pKa = 3.19YY176 pKa = 11.37GGKK179 pKa = 7.59QTYY182 pKa = 8.75KK183 pKa = 10.53APSMPNYY190 pKa = 9.15KK191 pKa = 10.05KK192 pKa = 10.97AMNVNIRR199 pKa = 11.84NINVKK204 pKa = 7.29TQWKK208 pKa = 7.81DD209 pKa = 3.12TGGGKK214 pKa = 10.11YY215 pKa = 10.33EE216 pKa = 4.14DD217 pKa = 3.64VKK219 pKa = 11.28EE220 pKa = 3.89NAILYY225 pKa = 9.01VVVNDD230 pKa = 3.71NTDD233 pKa = 3.47NTNMYY238 pKa = 8.08ATLFGNCRR246 pKa = 11.84AYY248 pKa = 10.51FYY250 pKa = 11.49
MM1 pKa = 7.59AVQSQKK7 pKa = 10.4RR8 pKa = 11.84KK9 pKa = 10.22YY10 pKa = 9.69MSATNWAKK18 pKa = 10.31KK19 pKa = 10.17RR20 pKa = 11.84KK21 pKa = 5.06TTTGRR26 pKa = 11.84TVSKK30 pKa = 10.05KK31 pKa = 8.12YY32 pKa = 7.44QWKK35 pKa = 10.13RR36 pKa = 11.84PVRR39 pKa = 11.84SNRR42 pKa = 11.84SVKK45 pKa = 10.6LKK47 pKa = 10.23MYY49 pKa = 10.74DD50 pKa = 3.68DD51 pKa = 4.07MLGASGVGSTISNDD65 pKa = 3.59GMITMLNNYY74 pKa = 7.83VQGIGDD80 pKa = 3.83SQRR83 pKa = 11.84SNNLTVTRR91 pKa = 11.84HH92 pKa = 5.78LKK94 pKa = 10.43FDD96 pKa = 3.68MALMASSMFWEE107 pKa = 4.88RR108 pKa = 11.84PNYY111 pKa = 5.2MTQYY115 pKa = 9.32HH116 pKa = 5.44WVIVDD121 pKa = 3.71RR122 pKa = 11.84DD123 pKa = 3.63VGATFPTQLSTIFDD137 pKa = 3.94IPTGGQAMPSTYY149 pKa = 10.12RR150 pKa = 11.84IRR152 pKa = 11.84RR153 pKa = 11.84DD154 pKa = 3.09ANEE157 pKa = 3.67RR158 pKa = 11.84FIVKK162 pKa = 10.08KK163 pKa = 10.17KK164 pKa = 8.49WQTHH168 pKa = 4.72LMSSGTDD175 pKa = 3.19YY176 pKa = 11.37GGKK179 pKa = 7.59QTYY182 pKa = 8.75KK183 pKa = 10.53APSMPNYY190 pKa = 9.15KK191 pKa = 10.05KK192 pKa = 10.97AMNVNIRR199 pKa = 11.84NINVKK204 pKa = 7.29TQWKK208 pKa = 7.81DD209 pKa = 3.12TGGGKK214 pKa = 10.11YY215 pKa = 10.33EE216 pKa = 4.14DD217 pKa = 3.64VKK219 pKa = 11.28EE220 pKa = 3.89NAILYY225 pKa = 9.01VVVNDD230 pKa = 3.71NTDD233 pKa = 3.47NTNMYY238 pKa = 8.08ATLFGNCRR246 pKa = 11.84AYY248 pKa = 10.51FYY250 pKa = 11.49
Molecular weight: 28.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
863 |
89 |
302 |
172.6 |
20.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.171 ± 0.806 | 1.97 ± 0.665 |
5.678 ± 0.837 | 6.373 ± 2.348 |
4.751 ± 0.933 | 5.214 ± 0.589 |
1.854 ± 0.516 | 5.678 ± 1.053 |
6.257 ± 1.409 | 7.648 ± 1.56 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.013 ± 1.237 | 5.446 ± 1.209 |
4.983 ± 0.898 | 4.403 ± 0.311 |
5.794 ± 1.094 | 7.764 ± 0.836 |
6.257 ± 1.315 | 5.562 ± 0.802 |
2.086 ± 0.527 | 5.098 ± 0.71 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
