
Sclerotinia sclerotiorum umbra-like virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Calvusvirinae; Umbravirus; unclassified Umbravirus
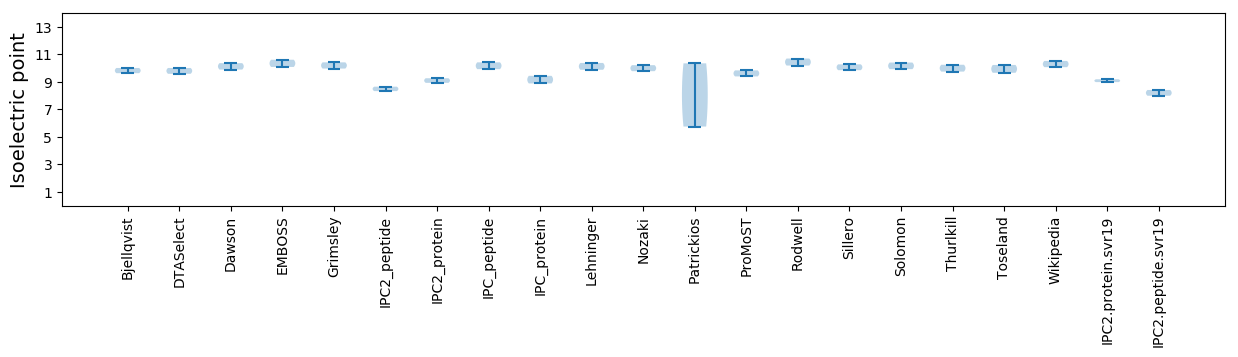
Average proteome isoelectric point is 9.09
Get precalculated fractions of proteins
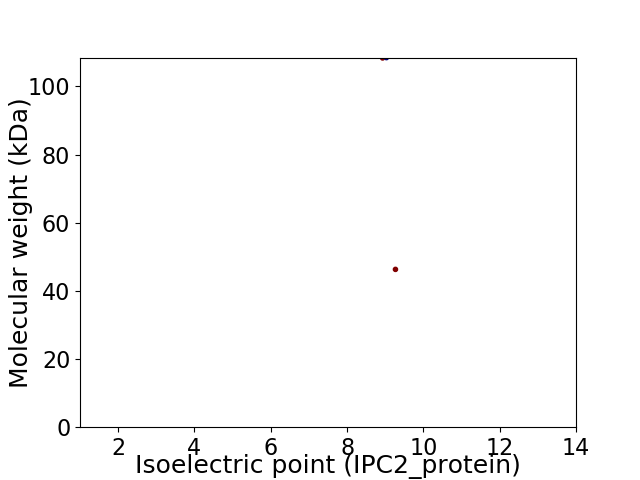
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A172T0R0|A0A172T0R0_9TOMB Replication-associated protein OS=Sclerotinia sclerotiorum umbra-like virus 1 OX=1435450 PE=4 SV=1
MM1 pKa = 7.16STDD4 pKa = 3.47TKK6 pKa = 9.98QTAHH10 pKa = 6.57RR11 pKa = 11.84CLRR14 pKa = 11.84LQPQAPPRR22 pKa = 11.84DD23 pKa = 3.43RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 10.79LSVGGLTTTVVDD38 pKa = 3.5WRR40 pKa = 11.84LAAHH44 pKa = 6.2PHH46 pKa = 5.26RR47 pKa = 11.84RR48 pKa = 11.84WWLTPNDD55 pKa = 3.0FMMFKK60 pKa = 10.74FLRR63 pKa = 11.84KK64 pKa = 9.71NKK66 pKa = 9.69IEE68 pKa = 4.24STTPDD73 pKa = 4.61PIEE76 pKa = 4.47SFAASAFSLDD86 pKa = 3.56HH87 pKa = 7.58DD88 pKa = 4.67EE89 pKa = 4.94GRR91 pKa = 11.84HH92 pKa = 4.42TPVVRR97 pKa = 11.84LPKK100 pKa = 9.84VVPPVEE106 pKa = 4.29YY107 pKa = 10.62VIKK110 pKa = 10.45DD111 pKa = 3.75YY112 pKa = 11.46LQGASGRR119 pKa = 11.84WDD121 pKa = 3.67VPYY124 pKa = 9.96MVYY127 pKa = 10.41PPLSLAGQPAPPDD140 pKa = 3.5VATSTDD146 pKa = 2.92VDD148 pKa = 3.81VGVVEE153 pKa = 5.0GEE155 pKa = 4.07VKK157 pKa = 10.55GRR159 pKa = 11.84VVVRR163 pKa = 11.84LPAAKK168 pKa = 9.4QVRR171 pKa = 11.84FDD173 pKa = 3.58GRR175 pKa = 11.84QTEE178 pKa = 4.06ASTGSASLEE187 pKa = 3.94KK188 pKa = 10.26KK189 pKa = 10.12KK190 pKa = 10.37PSRR193 pKa = 11.84EE194 pKa = 3.99FKK196 pKa = 10.51PSTKK200 pKa = 10.13YY201 pKa = 10.86GPSVLEE207 pKa = 3.91NEE209 pKa = 4.24RR210 pKa = 11.84GEE212 pKa = 4.46LRR214 pKa = 11.84DD215 pKa = 3.47AEE217 pKa = 4.32FGFRR221 pKa = 11.84LRR223 pKa = 11.84KK224 pKa = 9.56RR225 pKa = 11.84GRR227 pKa = 11.84LTRR230 pKa = 11.84LLEE233 pKa = 4.59SIAKK237 pKa = 9.75RR238 pKa = 11.84DD239 pKa = 3.55VSHH242 pKa = 7.05QSGEE246 pKa = 4.09GGKK249 pKa = 8.49TSEE252 pKa = 3.89QPARR256 pKa = 11.84HH257 pKa = 5.46WLGLITRR264 pKa = 11.84WGGKK268 pKa = 7.97LTVTQRR274 pKa = 11.84VKK276 pKa = 11.22LNNTDD281 pKa = 3.21KK282 pKa = 11.36DD283 pKa = 4.12EE284 pKa = 4.63PSEE287 pKa = 4.09DD288 pKa = 3.47CYY290 pKa = 9.51MTVRR294 pKa = 11.84IGGSVLAASARR305 pKa = 11.84LLAGLVVSATNKK317 pKa = 9.41RR318 pKa = 11.84RR319 pKa = 11.84TRR321 pKa = 11.84EE322 pKa = 3.86LQLALTAKK330 pKa = 10.09ARR332 pKa = 11.84QIATASGMTDD342 pKa = 2.5SYY344 pKa = 11.52LAMVLAGTVALAMRR358 pKa = 11.84VSGPEE363 pKa = 3.53QMANEE368 pKa = 4.22SLGGVSGRR376 pKa = 11.84QSQRR380 pKa = 11.84ASEE383 pKa = 4.92DD384 pKa = 2.65IHH386 pKa = 8.55AGRR389 pKa = 11.84TTSLFRR395 pKa = 11.84QFTRR399 pKa = 11.84AVGDD403 pKa = 3.5VMRR406 pKa = 11.84VEE408 pKa = 4.62CVTGVVLPRR417 pKa = 11.84ATTXWGVSNSGIGVCVGSKK436 pKa = 8.22EE437 pKa = 3.93QHH439 pKa = 6.98KK440 pKa = 9.91IRR442 pKa = 11.84QGARR446 pKa = 11.84MVIPSLDD453 pKa = 3.8TTQHH457 pKa = 5.11SCGPEE462 pKa = 3.41ARR464 pKa = 11.84RR465 pKa = 11.84RR466 pKa = 11.84MYY468 pKa = 10.22RR469 pKa = 11.84VWVPSVDD476 pKa = 4.12GLWAPAVHH484 pKa = 6.91ANCVHH489 pKa = 6.31NEE491 pKa = 3.62EE492 pKa = 4.11AALRR496 pKa = 11.84LRR498 pKa = 11.84TLGPTPADD506 pKa = 3.52PVCPEE511 pKa = 4.12LDD513 pKa = 3.21KK514 pKa = 11.66AFARR518 pKa = 11.84VGVLLSRR525 pKa = 11.84LNIQRR530 pKa = 11.84WTKK533 pKa = 7.95QQVVEE538 pKa = 4.58SYY540 pKa = 10.04SGRR543 pKa = 11.84MRR545 pKa = 11.84QRR547 pKa = 11.84YY548 pKa = 7.14QAALEE553 pKa = 4.22SLVEE557 pKa = 4.09EE558 pKa = 4.68PRR560 pKa = 11.84LSKK563 pKa = 10.56YY564 pKa = 9.81DD565 pKa = 3.41RR566 pKa = 11.84RR567 pKa = 11.84LTGFLKK573 pKa = 10.78AEE575 pKa = 4.3KK576 pKa = 10.08FNPLLKK582 pKa = 10.0QSKK585 pKa = 9.34PRR587 pKa = 11.84MIMCRR592 pKa = 11.84SPRR595 pKa = 11.84FNLEE599 pKa = 3.51LASYY603 pKa = 8.93LKK605 pKa = 10.34PLEE608 pKa = 4.27HH609 pKa = 6.82ALWRR613 pKa = 11.84RR614 pKa = 11.84WKK616 pKa = 9.49YY617 pKa = 10.74GRR619 pKa = 11.84GGVTPTRR626 pKa = 11.84VVGKK630 pKa = 9.87GLNGEE635 pKa = 4.12QRR637 pKa = 11.84ASLIRR642 pKa = 11.84RR643 pKa = 11.84KK644 pKa = 9.07MEE646 pKa = 4.15SVGEE650 pKa = 4.21CTVFEE655 pKa = 4.31IDD657 pKa = 3.21GKK659 pKa = 10.9AFEE662 pKa = 4.22AHH664 pKa = 3.99VTLRR668 pKa = 11.84QVDD671 pKa = 4.52LEE673 pKa = 4.27HH674 pKa = 6.91KK675 pKa = 9.74VYY677 pKa = 10.87KK678 pKa = 10.1KK679 pKa = 10.66VYY681 pKa = 8.42PRR683 pKa = 11.84DD684 pKa = 3.61ARR686 pKa = 11.84LHH688 pKa = 7.04DD689 pKa = 4.3LLQTQRR695 pKa = 11.84KK696 pKa = 8.86LKK698 pKa = 10.31GKK700 pKa = 7.22TLGGIKK706 pKa = 9.97FEE708 pKa = 4.73RR709 pKa = 11.84EE710 pKa = 3.83GCRR713 pKa = 11.84ASGDD717 pKa = 3.86FNTGLGNTLVMGSCVDD733 pKa = 4.29AILRR737 pKa = 11.84SAEE740 pKa = 4.03SALGAFRR747 pKa = 11.84TTFLADD753 pKa = 3.26GDD755 pKa = 4.02NALVFVEE762 pKa = 4.43TSKK765 pKa = 11.66AEE767 pKa = 4.04GVRR770 pKa = 11.84SFFSSMVHH778 pKa = 4.44TVVGQEE784 pKa = 3.95LTVEE788 pKa = 4.27KK789 pKa = 8.87PTTIYY794 pKa = 10.87EE795 pKa = 4.12EE796 pKa = 4.67VVFGQCKK803 pKa = 8.76PCFNGDD809 pKa = 3.22HH810 pKa = 5.59YY811 pKa = 10.27TMVRR815 pKa = 11.84HH816 pKa = 5.9PLKK819 pKa = 9.78TLSNAFSGYY828 pKa = 7.93RR829 pKa = 11.84HH830 pKa = 6.47YY831 pKa = 11.13DD832 pKa = 3.0QSAYY836 pKa = 10.62VGPALKK842 pKa = 10.18AVCRR846 pKa = 11.84AEE848 pKa = 4.18LALSRR853 pKa = 11.84GIPLLEE859 pKa = 4.56AYY861 pKa = 8.7FARR864 pKa = 11.84ALEE867 pKa = 4.03RR868 pKa = 11.84LEE870 pKa = 4.17VYY872 pKa = 10.42RR873 pKa = 11.84DD874 pKa = 3.48LKK876 pKa = 11.12DD877 pKa = 3.49PSVVLEE883 pKa = 3.93DD884 pKa = 3.91RR885 pKa = 11.84LRR887 pKa = 11.84HH888 pKa = 5.43ALAPAYY894 pKa = 9.51AAKK897 pKa = 10.37SSNRR901 pKa = 11.84GVGMDD906 pKa = 4.23ARR908 pKa = 11.84LSFQKK913 pKa = 10.46AWGIGVEE920 pKa = 4.23EE921 pKa = 4.33QILMEE926 pKa = 4.67AEE928 pKa = 4.05LVASVDD934 pKa = 3.04HH935 pKa = 6.91WDD937 pKa = 3.23WTTLRR942 pKa = 11.84EE943 pKa = 4.12DD944 pKa = 3.48VHH946 pKa = 8.37VGDD949 pKa = 5.68GPFADD954 pKa = 3.68TDD956 pKa = 4.01LVEE959 pKa = 4.2NRR961 pKa = 11.84VDD963 pKa = 3.67LFLSGRR969 pKa = 11.84RR970 pKa = 11.84LKK972 pKa = 10.83DD973 pKa = 3.02
MM1 pKa = 7.16STDD4 pKa = 3.47TKK6 pKa = 9.98QTAHH10 pKa = 6.57RR11 pKa = 11.84CLRR14 pKa = 11.84LQPQAPPRR22 pKa = 11.84DD23 pKa = 3.43RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 10.79LSVGGLTTTVVDD38 pKa = 3.5WRR40 pKa = 11.84LAAHH44 pKa = 6.2PHH46 pKa = 5.26RR47 pKa = 11.84RR48 pKa = 11.84WWLTPNDD55 pKa = 3.0FMMFKK60 pKa = 10.74FLRR63 pKa = 11.84KK64 pKa = 9.71NKK66 pKa = 9.69IEE68 pKa = 4.24STTPDD73 pKa = 4.61PIEE76 pKa = 4.47SFAASAFSLDD86 pKa = 3.56HH87 pKa = 7.58DD88 pKa = 4.67EE89 pKa = 4.94GRR91 pKa = 11.84HH92 pKa = 4.42TPVVRR97 pKa = 11.84LPKK100 pKa = 9.84VVPPVEE106 pKa = 4.29YY107 pKa = 10.62VIKK110 pKa = 10.45DD111 pKa = 3.75YY112 pKa = 11.46LQGASGRR119 pKa = 11.84WDD121 pKa = 3.67VPYY124 pKa = 9.96MVYY127 pKa = 10.41PPLSLAGQPAPPDD140 pKa = 3.5VATSTDD146 pKa = 2.92VDD148 pKa = 3.81VGVVEE153 pKa = 5.0GEE155 pKa = 4.07VKK157 pKa = 10.55GRR159 pKa = 11.84VVVRR163 pKa = 11.84LPAAKK168 pKa = 9.4QVRR171 pKa = 11.84FDD173 pKa = 3.58GRR175 pKa = 11.84QTEE178 pKa = 4.06ASTGSASLEE187 pKa = 3.94KK188 pKa = 10.26KK189 pKa = 10.12KK190 pKa = 10.37PSRR193 pKa = 11.84EE194 pKa = 3.99FKK196 pKa = 10.51PSTKK200 pKa = 10.13YY201 pKa = 10.86GPSVLEE207 pKa = 3.91NEE209 pKa = 4.24RR210 pKa = 11.84GEE212 pKa = 4.46LRR214 pKa = 11.84DD215 pKa = 3.47AEE217 pKa = 4.32FGFRR221 pKa = 11.84LRR223 pKa = 11.84KK224 pKa = 9.56RR225 pKa = 11.84GRR227 pKa = 11.84LTRR230 pKa = 11.84LLEE233 pKa = 4.59SIAKK237 pKa = 9.75RR238 pKa = 11.84DD239 pKa = 3.55VSHH242 pKa = 7.05QSGEE246 pKa = 4.09GGKK249 pKa = 8.49TSEE252 pKa = 3.89QPARR256 pKa = 11.84HH257 pKa = 5.46WLGLITRR264 pKa = 11.84WGGKK268 pKa = 7.97LTVTQRR274 pKa = 11.84VKK276 pKa = 11.22LNNTDD281 pKa = 3.21KK282 pKa = 11.36DD283 pKa = 4.12EE284 pKa = 4.63PSEE287 pKa = 4.09DD288 pKa = 3.47CYY290 pKa = 9.51MTVRR294 pKa = 11.84IGGSVLAASARR305 pKa = 11.84LLAGLVVSATNKK317 pKa = 9.41RR318 pKa = 11.84RR319 pKa = 11.84TRR321 pKa = 11.84EE322 pKa = 3.86LQLALTAKK330 pKa = 10.09ARR332 pKa = 11.84QIATASGMTDD342 pKa = 2.5SYY344 pKa = 11.52LAMVLAGTVALAMRR358 pKa = 11.84VSGPEE363 pKa = 3.53QMANEE368 pKa = 4.22SLGGVSGRR376 pKa = 11.84QSQRR380 pKa = 11.84ASEE383 pKa = 4.92DD384 pKa = 2.65IHH386 pKa = 8.55AGRR389 pKa = 11.84TTSLFRR395 pKa = 11.84QFTRR399 pKa = 11.84AVGDD403 pKa = 3.5VMRR406 pKa = 11.84VEE408 pKa = 4.62CVTGVVLPRR417 pKa = 11.84ATTXWGVSNSGIGVCVGSKK436 pKa = 8.22EE437 pKa = 3.93QHH439 pKa = 6.98KK440 pKa = 9.91IRR442 pKa = 11.84QGARR446 pKa = 11.84MVIPSLDD453 pKa = 3.8TTQHH457 pKa = 5.11SCGPEE462 pKa = 3.41ARR464 pKa = 11.84RR465 pKa = 11.84RR466 pKa = 11.84MYY468 pKa = 10.22RR469 pKa = 11.84VWVPSVDD476 pKa = 4.12GLWAPAVHH484 pKa = 6.91ANCVHH489 pKa = 6.31NEE491 pKa = 3.62EE492 pKa = 4.11AALRR496 pKa = 11.84LRR498 pKa = 11.84TLGPTPADD506 pKa = 3.52PVCPEE511 pKa = 4.12LDD513 pKa = 3.21KK514 pKa = 11.66AFARR518 pKa = 11.84VGVLLSRR525 pKa = 11.84LNIQRR530 pKa = 11.84WTKK533 pKa = 7.95QQVVEE538 pKa = 4.58SYY540 pKa = 10.04SGRR543 pKa = 11.84MRR545 pKa = 11.84QRR547 pKa = 11.84YY548 pKa = 7.14QAALEE553 pKa = 4.22SLVEE557 pKa = 4.09EE558 pKa = 4.68PRR560 pKa = 11.84LSKK563 pKa = 10.56YY564 pKa = 9.81DD565 pKa = 3.41RR566 pKa = 11.84RR567 pKa = 11.84LTGFLKK573 pKa = 10.78AEE575 pKa = 4.3KK576 pKa = 10.08FNPLLKK582 pKa = 10.0QSKK585 pKa = 9.34PRR587 pKa = 11.84MIMCRR592 pKa = 11.84SPRR595 pKa = 11.84FNLEE599 pKa = 3.51LASYY603 pKa = 8.93LKK605 pKa = 10.34PLEE608 pKa = 4.27HH609 pKa = 6.82ALWRR613 pKa = 11.84RR614 pKa = 11.84WKK616 pKa = 9.49YY617 pKa = 10.74GRR619 pKa = 11.84GGVTPTRR626 pKa = 11.84VVGKK630 pKa = 9.87GLNGEE635 pKa = 4.12QRR637 pKa = 11.84ASLIRR642 pKa = 11.84RR643 pKa = 11.84KK644 pKa = 9.07MEE646 pKa = 4.15SVGEE650 pKa = 4.21CTVFEE655 pKa = 4.31IDD657 pKa = 3.21GKK659 pKa = 10.9AFEE662 pKa = 4.22AHH664 pKa = 3.99VTLRR668 pKa = 11.84QVDD671 pKa = 4.52LEE673 pKa = 4.27HH674 pKa = 6.91KK675 pKa = 9.74VYY677 pKa = 10.87KK678 pKa = 10.1KK679 pKa = 10.66VYY681 pKa = 8.42PRR683 pKa = 11.84DD684 pKa = 3.61ARR686 pKa = 11.84LHH688 pKa = 7.04DD689 pKa = 4.3LLQTQRR695 pKa = 11.84KK696 pKa = 8.86LKK698 pKa = 10.31GKK700 pKa = 7.22TLGGIKK706 pKa = 9.97FEE708 pKa = 4.73RR709 pKa = 11.84EE710 pKa = 3.83GCRR713 pKa = 11.84ASGDD717 pKa = 3.86FNTGLGNTLVMGSCVDD733 pKa = 4.29AILRR737 pKa = 11.84SAEE740 pKa = 4.03SALGAFRR747 pKa = 11.84TTFLADD753 pKa = 3.26GDD755 pKa = 4.02NALVFVEE762 pKa = 4.43TSKK765 pKa = 11.66AEE767 pKa = 4.04GVRR770 pKa = 11.84SFFSSMVHH778 pKa = 4.44TVVGQEE784 pKa = 3.95LTVEE788 pKa = 4.27KK789 pKa = 8.87PTTIYY794 pKa = 10.87EE795 pKa = 4.12EE796 pKa = 4.67VVFGQCKK803 pKa = 8.76PCFNGDD809 pKa = 3.22HH810 pKa = 5.59YY811 pKa = 10.27TMVRR815 pKa = 11.84HH816 pKa = 5.9PLKK819 pKa = 9.78TLSNAFSGYY828 pKa = 7.93RR829 pKa = 11.84HH830 pKa = 6.47YY831 pKa = 11.13DD832 pKa = 3.0QSAYY836 pKa = 10.62VGPALKK842 pKa = 10.18AVCRR846 pKa = 11.84AEE848 pKa = 4.18LALSRR853 pKa = 11.84GIPLLEE859 pKa = 4.56AYY861 pKa = 8.7FARR864 pKa = 11.84ALEE867 pKa = 4.03RR868 pKa = 11.84LEE870 pKa = 4.17VYY872 pKa = 10.42RR873 pKa = 11.84DD874 pKa = 3.48LKK876 pKa = 11.12DD877 pKa = 3.49PSVVLEE883 pKa = 3.93DD884 pKa = 3.91RR885 pKa = 11.84LRR887 pKa = 11.84HH888 pKa = 5.43ALAPAYY894 pKa = 9.51AAKK897 pKa = 10.37SSNRR901 pKa = 11.84GVGMDD906 pKa = 4.23ARR908 pKa = 11.84LSFQKK913 pKa = 10.46AWGIGVEE920 pKa = 4.23EE921 pKa = 4.33QILMEE926 pKa = 4.67AEE928 pKa = 4.05LVASVDD934 pKa = 3.04HH935 pKa = 6.91WDD937 pKa = 3.23WTTLRR942 pKa = 11.84EE943 pKa = 4.12DD944 pKa = 3.48VHH946 pKa = 8.37VGDD949 pKa = 5.68GPFADD954 pKa = 3.68TDD956 pKa = 4.01LVEE959 pKa = 4.2NRR961 pKa = 11.84VDD963 pKa = 3.67LFLSGRR969 pKa = 11.84RR970 pKa = 11.84LKK972 pKa = 10.83DD973 pKa = 3.02
Molecular weight: 108.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A172T0R0|A0A172T0R0_9TOMB Replication-associated protein OS=Sclerotinia sclerotiorum umbra-like virus 1 OX=1435450 PE=4 SV=1
MM1 pKa = 7.16STDD4 pKa = 3.47TKK6 pKa = 9.98QTAHH10 pKa = 6.57RR11 pKa = 11.84CLRR14 pKa = 11.84LQPQAPPRR22 pKa = 11.84DD23 pKa = 3.43RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 10.79LSVGGLTTTVVDD38 pKa = 3.5WRR40 pKa = 11.84LAAHH44 pKa = 6.2PHH46 pKa = 5.26RR47 pKa = 11.84RR48 pKa = 11.84WWLTPNDD55 pKa = 3.0FMMFKK60 pKa = 10.74FLRR63 pKa = 11.84KK64 pKa = 9.71NKK66 pKa = 9.69IEE68 pKa = 4.24STTPDD73 pKa = 4.61PIEE76 pKa = 4.47SFAASAFSLDD86 pKa = 3.56HH87 pKa = 7.58DD88 pKa = 4.67EE89 pKa = 4.94GRR91 pKa = 11.84HH92 pKa = 4.42TPVVRR97 pKa = 11.84LPKK100 pKa = 9.84VVPPVEE106 pKa = 4.29YY107 pKa = 10.62VIKK110 pKa = 10.45DD111 pKa = 3.75YY112 pKa = 11.46LQGASGRR119 pKa = 11.84WDD121 pKa = 3.67VPYY124 pKa = 9.96MVYY127 pKa = 10.41PPLSLAGQPAPPDD140 pKa = 3.5VATSTDD146 pKa = 2.92VDD148 pKa = 3.81VGVVEE153 pKa = 5.0GEE155 pKa = 4.07VKK157 pKa = 10.55GRR159 pKa = 11.84VVVRR163 pKa = 11.84LPAAKK168 pKa = 9.4QVRR171 pKa = 11.84FDD173 pKa = 3.58GRR175 pKa = 11.84QTEE178 pKa = 4.06ASTGSASLEE187 pKa = 3.94KK188 pKa = 10.26KK189 pKa = 10.12KK190 pKa = 10.37PSRR193 pKa = 11.84EE194 pKa = 3.99FKK196 pKa = 10.51PSTKK200 pKa = 10.13YY201 pKa = 10.86GPSVLEE207 pKa = 3.91NEE209 pKa = 4.24RR210 pKa = 11.84GEE212 pKa = 4.46LRR214 pKa = 11.84DD215 pKa = 3.47AEE217 pKa = 4.32FGFRR221 pKa = 11.84LRR223 pKa = 11.84KK224 pKa = 9.56RR225 pKa = 11.84GRR227 pKa = 11.84LTRR230 pKa = 11.84LLEE233 pKa = 4.59SIAKK237 pKa = 9.75RR238 pKa = 11.84DD239 pKa = 3.55VSHH242 pKa = 7.05QSGEE246 pKa = 4.09GGKK249 pKa = 8.49TSEE252 pKa = 3.89QPARR256 pKa = 11.84HH257 pKa = 5.46WLGLITRR264 pKa = 11.84WGGKK268 pKa = 7.97LTVTQRR274 pKa = 11.84VKK276 pKa = 11.22LNNTDD281 pKa = 3.21KK282 pKa = 11.36DD283 pKa = 4.12EE284 pKa = 4.63PSEE287 pKa = 4.09DD288 pKa = 3.47CYY290 pKa = 9.51MTVRR294 pKa = 11.84IGGSVLAASARR305 pKa = 11.84LLAGLVVSATNKK317 pKa = 9.41RR318 pKa = 11.84RR319 pKa = 11.84TRR321 pKa = 11.84EE322 pKa = 3.86LQLALTAKK330 pKa = 10.09ARR332 pKa = 11.84QIATASGMTDD342 pKa = 2.5SYY344 pKa = 11.52LAMVLAGTVALAMRR358 pKa = 11.84VSGPEE363 pKa = 3.53QMANEE368 pKa = 4.22SLGGVSGRR376 pKa = 11.84QSQRR380 pKa = 11.84ASEE383 pKa = 4.92DD384 pKa = 2.65IHH386 pKa = 8.55AGRR389 pKa = 11.84TTSLFRR395 pKa = 11.84QFTRR399 pKa = 11.84AVGDD403 pKa = 3.5VMRR406 pKa = 11.84VEE408 pKa = 4.62CVTGVVLPRR417 pKa = 11.84ATTT420 pKa = 3.33
MM1 pKa = 7.16STDD4 pKa = 3.47TKK6 pKa = 9.98QTAHH10 pKa = 6.57RR11 pKa = 11.84CLRR14 pKa = 11.84LQPQAPPRR22 pKa = 11.84DD23 pKa = 3.43RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 10.79LSVGGLTTTVVDD38 pKa = 3.5WRR40 pKa = 11.84LAAHH44 pKa = 6.2PHH46 pKa = 5.26RR47 pKa = 11.84RR48 pKa = 11.84WWLTPNDD55 pKa = 3.0FMMFKK60 pKa = 10.74FLRR63 pKa = 11.84KK64 pKa = 9.71NKK66 pKa = 9.69IEE68 pKa = 4.24STTPDD73 pKa = 4.61PIEE76 pKa = 4.47SFAASAFSLDD86 pKa = 3.56HH87 pKa = 7.58DD88 pKa = 4.67EE89 pKa = 4.94GRR91 pKa = 11.84HH92 pKa = 4.42TPVVRR97 pKa = 11.84LPKK100 pKa = 9.84VVPPVEE106 pKa = 4.29YY107 pKa = 10.62VIKK110 pKa = 10.45DD111 pKa = 3.75YY112 pKa = 11.46LQGASGRR119 pKa = 11.84WDD121 pKa = 3.67VPYY124 pKa = 9.96MVYY127 pKa = 10.41PPLSLAGQPAPPDD140 pKa = 3.5VATSTDD146 pKa = 2.92VDD148 pKa = 3.81VGVVEE153 pKa = 5.0GEE155 pKa = 4.07VKK157 pKa = 10.55GRR159 pKa = 11.84VVVRR163 pKa = 11.84LPAAKK168 pKa = 9.4QVRR171 pKa = 11.84FDD173 pKa = 3.58GRR175 pKa = 11.84QTEE178 pKa = 4.06ASTGSASLEE187 pKa = 3.94KK188 pKa = 10.26KK189 pKa = 10.12KK190 pKa = 10.37PSRR193 pKa = 11.84EE194 pKa = 3.99FKK196 pKa = 10.51PSTKK200 pKa = 10.13YY201 pKa = 10.86GPSVLEE207 pKa = 3.91NEE209 pKa = 4.24RR210 pKa = 11.84GEE212 pKa = 4.46LRR214 pKa = 11.84DD215 pKa = 3.47AEE217 pKa = 4.32FGFRR221 pKa = 11.84LRR223 pKa = 11.84KK224 pKa = 9.56RR225 pKa = 11.84GRR227 pKa = 11.84LTRR230 pKa = 11.84LLEE233 pKa = 4.59SIAKK237 pKa = 9.75RR238 pKa = 11.84DD239 pKa = 3.55VSHH242 pKa = 7.05QSGEE246 pKa = 4.09GGKK249 pKa = 8.49TSEE252 pKa = 3.89QPARR256 pKa = 11.84HH257 pKa = 5.46WLGLITRR264 pKa = 11.84WGGKK268 pKa = 7.97LTVTQRR274 pKa = 11.84VKK276 pKa = 11.22LNNTDD281 pKa = 3.21KK282 pKa = 11.36DD283 pKa = 4.12EE284 pKa = 4.63PSEE287 pKa = 4.09DD288 pKa = 3.47CYY290 pKa = 9.51MTVRR294 pKa = 11.84IGGSVLAASARR305 pKa = 11.84LLAGLVVSATNKK317 pKa = 9.41RR318 pKa = 11.84RR319 pKa = 11.84TRR321 pKa = 11.84EE322 pKa = 3.86LQLALTAKK330 pKa = 10.09ARR332 pKa = 11.84QIATASGMTDD342 pKa = 2.5SYY344 pKa = 11.52LAMVLAGTVALAMRR358 pKa = 11.84VSGPEE363 pKa = 3.53QMANEE368 pKa = 4.22SLGGVSGRR376 pKa = 11.84QSQRR380 pKa = 11.84ASEE383 pKa = 4.92DD384 pKa = 2.65IHH386 pKa = 8.55AGRR389 pKa = 11.84TTSLFRR395 pKa = 11.84QFTRR399 pKa = 11.84AVGDD403 pKa = 3.5VMRR406 pKa = 11.84VEE408 pKa = 4.62CVTGVVLPRR417 pKa = 11.84ATTT420 pKa = 3.33
Molecular weight: 46.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1393 |
420 |
973 |
696.5 |
77.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.615 ± 0.107 | 1.22 ± 0.278 |
4.953 ± 0.026 | 6.03 ± 0.304 |
3.015 ± 0.217 | 7.825 ± 0.018 |
2.225 ± 0.176 | 2.082 ± 0.097 |
5.169 ± 0.093 | 9.476 ± 0.366 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.225 ± 0.085 | 2.01 ± 0.188 |
5.24 ± 0.39 | 3.589 ± 0.121 |
9.691 ± 0.169 | 6.892 ± 0.268 |
6.748 ± 0.739 | 9.117 ± 0.093 |
1.508 ± 0.043 | 2.297 ± 0.215 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
