
Clostridium sp. CAG:242
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
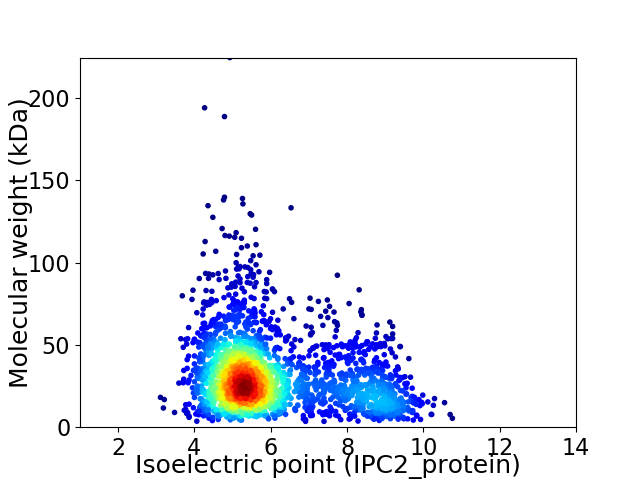
Virtual 2D-PAGE plot for 2433 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
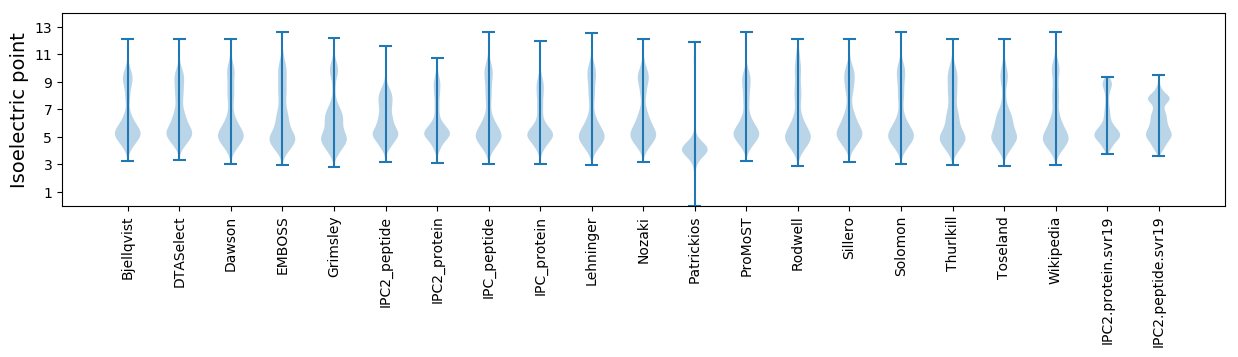
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6CXA1|R6CXA1_9CLOT UPF0272 protein Mahau_1643 OS=Clostridium sp. CAG:242 OX=1262783 GN=BN558_01493 PE=4 SV=1
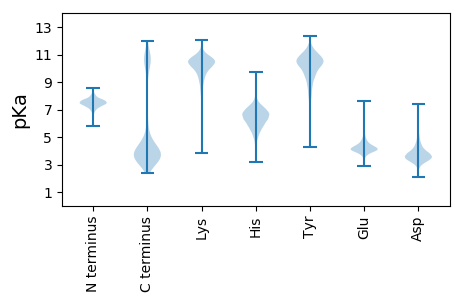
MM1 pKa = 7.32KK2 pKa = 10.48RR3 pKa = 11.84MFILIFVAVLLFSLAACGNNEE24 pKa = 4.13GTFSDD29 pKa = 3.52QTINNNLSQSEE40 pKa = 4.4GVDD43 pKa = 3.22NSQVQQIHH51 pKa = 4.96TNILIAYY58 pKa = 8.99DD59 pKa = 3.97PEE61 pKa = 4.25QEE63 pKa = 4.44TITRR67 pKa = 11.84AAKK70 pKa = 10.48LLADD74 pKa = 4.35TLNADD79 pKa = 5.2LIEE82 pKa = 4.33ITNDD86 pKa = 2.78SNLQVDD92 pKa = 4.2TYY94 pKa = 11.32EE95 pKa = 4.17FVLLGFDD102 pKa = 3.91GHH104 pKa = 5.83NQQLSPQVQNFLKK117 pKa = 10.25RR118 pKa = 11.84YY119 pKa = 8.32NFGARR124 pKa = 11.84TIYY127 pKa = 10.39PFVLGDD133 pKa = 4.59GIDD136 pKa = 3.82LSAVSSAISEE146 pKa = 4.41LEE148 pKa = 3.87PGALMGNGEE157 pKa = 4.32FLIEE161 pKa = 4.17EE162 pKa = 4.25NTTDD166 pKa = 3.46EE167 pKa = 5.49EE168 pKa = 4.07IRR170 pKa = 11.84EE171 pKa = 4.23WGAEE175 pKa = 4.08LEE177 pKa = 4.21LSDD180 pKa = 4.89IKK182 pKa = 10.35TAVPTAEE189 pKa = 4.43EE190 pKa = 4.83GNTVATATVTPDD202 pKa = 3.3QQQVLYY208 pKa = 10.54LWEE211 pKa = 4.43EE212 pKa = 4.45DD213 pKa = 3.39NAPAQTEE220 pKa = 4.12YY221 pKa = 11.09TMNDD225 pKa = 2.48GRR227 pKa = 11.84YY228 pKa = 9.63SDD230 pKa = 4.95NPDD233 pKa = 3.07FRR235 pKa = 11.84PYY237 pKa = 8.94LTSYY241 pKa = 9.47PVPEE245 pKa = 4.47GMDD248 pKa = 2.8IKK250 pKa = 10.84GAVLICPGGAFQFRR264 pKa = 11.84SDD266 pKa = 3.47QPEE269 pKa = 4.16GVDD272 pKa = 3.35VAEE275 pKa = 4.7ALNILGYY282 pKa = 9.82QCFVVDD288 pKa = 5.1YY289 pKa = 9.91RR290 pKa = 11.84LQPYY294 pKa = 7.25TQQEE298 pKa = 4.39GALDD302 pKa = 3.87LARR305 pKa = 11.84AVRR308 pKa = 11.84FVRR311 pKa = 11.84SHH313 pKa = 6.85AEE315 pKa = 3.75EE316 pKa = 4.41YY317 pKa = 10.8GIEE320 pKa = 4.15PQDD323 pKa = 3.33IAVMGFSAGGILSGEE338 pKa = 3.95MLINFDD344 pKa = 4.19GDD346 pKa = 3.88VLPTILDD353 pKa = 3.38PEE355 pKa = 4.35YY356 pKa = 10.85QQDD359 pKa = 3.78EE360 pKa = 4.65LDD362 pKa = 3.91SVSADD367 pKa = 3.17VAACGMIYY375 pKa = 10.34SFYY378 pKa = 10.69GQLSLGTTDD387 pKa = 3.56VDD389 pKa = 4.22LLRR392 pKa = 11.84SGDD395 pKa = 4.0LPPTFYY401 pKa = 10.84CYY403 pKa = 9.14GTRR406 pKa = 11.84DD407 pKa = 3.44PFYY410 pKa = 11.23GQFLANADD418 pKa = 3.62AAEE421 pKa = 4.05QAGVLVEE428 pKa = 4.78QLQLDD433 pKa = 4.07NMPHH437 pKa = 6.36GFGAGGDD444 pKa = 3.89WIPAYY449 pKa = 10.22DD450 pKa = 3.36KK451 pKa = 10.32WLTEE455 pKa = 4.06IFSQQ459 pKa = 4.81
MM1 pKa = 7.32KK2 pKa = 10.48RR3 pKa = 11.84MFILIFVAVLLFSLAACGNNEE24 pKa = 4.13GTFSDD29 pKa = 3.52QTINNNLSQSEE40 pKa = 4.4GVDD43 pKa = 3.22NSQVQQIHH51 pKa = 4.96TNILIAYY58 pKa = 8.99DD59 pKa = 3.97PEE61 pKa = 4.25QEE63 pKa = 4.44TITRR67 pKa = 11.84AAKK70 pKa = 10.48LLADD74 pKa = 4.35TLNADD79 pKa = 5.2LIEE82 pKa = 4.33ITNDD86 pKa = 2.78SNLQVDD92 pKa = 4.2TYY94 pKa = 11.32EE95 pKa = 4.17FVLLGFDD102 pKa = 3.91GHH104 pKa = 5.83NQQLSPQVQNFLKK117 pKa = 10.25RR118 pKa = 11.84YY119 pKa = 8.32NFGARR124 pKa = 11.84TIYY127 pKa = 10.39PFVLGDD133 pKa = 4.59GIDD136 pKa = 3.82LSAVSSAISEE146 pKa = 4.41LEE148 pKa = 3.87PGALMGNGEE157 pKa = 4.32FLIEE161 pKa = 4.17EE162 pKa = 4.25NTTDD166 pKa = 3.46EE167 pKa = 5.49EE168 pKa = 4.07IRR170 pKa = 11.84EE171 pKa = 4.23WGAEE175 pKa = 4.08LEE177 pKa = 4.21LSDD180 pKa = 4.89IKK182 pKa = 10.35TAVPTAEE189 pKa = 4.43EE190 pKa = 4.83GNTVATATVTPDD202 pKa = 3.3QQQVLYY208 pKa = 10.54LWEE211 pKa = 4.43EE212 pKa = 4.45DD213 pKa = 3.39NAPAQTEE220 pKa = 4.12YY221 pKa = 11.09TMNDD225 pKa = 2.48GRR227 pKa = 11.84YY228 pKa = 9.63SDD230 pKa = 4.95NPDD233 pKa = 3.07FRR235 pKa = 11.84PYY237 pKa = 8.94LTSYY241 pKa = 9.47PVPEE245 pKa = 4.47GMDD248 pKa = 2.8IKK250 pKa = 10.84GAVLICPGGAFQFRR264 pKa = 11.84SDD266 pKa = 3.47QPEE269 pKa = 4.16GVDD272 pKa = 3.35VAEE275 pKa = 4.7ALNILGYY282 pKa = 9.82QCFVVDD288 pKa = 5.1YY289 pKa = 9.91RR290 pKa = 11.84LQPYY294 pKa = 7.25TQQEE298 pKa = 4.39GALDD302 pKa = 3.87LARR305 pKa = 11.84AVRR308 pKa = 11.84FVRR311 pKa = 11.84SHH313 pKa = 6.85AEE315 pKa = 3.75EE316 pKa = 4.41YY317 pKa = 10.8GIEE320 pKa = 4.15PQDD323 pKa = 3.33IAVMGFSAGGILSGEE338 pKa = 3.95MLINFDD344 pKa = 4.19GDD346 pKa = 3.88VLPTILDD353 pKa = 3.38PEE355 pKa = 4.35YY356 pKa = 10.85QQDD359 pKa = 3.78EE360 pKa = 4.65LDD362 pKa = 3.91SVSADD367 pKa = 3.17VAACGMIYY375 pKa = 10.34SFYY378 pKa = 10.69GQLSLGTTDD387 pKa = 3.56VDD389 pKa = 4.22LLRR392 pKa = 11.84SGDD395 pKa = 4.0LPPTFYY401 pKa = 10.84CYY403 pKa = 9.14GTRR406 pKa = 11.84DD407 pKa = 3.44PFYY410 pKa = 11.23GQFLANADD418 pKa = 3.62AAEE421 pKa = 4.05QAGVLVEE428 pKa = 4.78QLQLDD433 pKa = 4.07NMPHH437 pKa = 6.36GFGAGGDD444 pKa = 3.89WIPAYY449 pKa = 10.22DD450 pKa = 3.36KK451 pKa = 10.32WLTEE455 pKa = 4.06IFSQQ459 pKa = 4.81
Molecular weight: 50.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6CVA1|R6CVA1_9CLOT Uncharacterized protein OS=Clostridium sp. CAG:242 OX=1262783 GN=BN558_00653 PE=3 SV=1
MM1 pKa = 7.34EE2 pKa = 5.09KK3 pKa = 10.53EE4 pKa = 3.55RR5 pKa = 11.84TEE7 pKa = 4.12RR8 pKa = 11.84NDD10 pKa = 2.85RR11 pKa = 11.84RR12 pKa = 11.84NRR14 pKa = 11.84KK15 pKa = 8.07SRR17 pKa = 11.84KK18 pKa = 8.8KK19 pKa = 10.01VCAFCMDD26 pKa = 4.14KK27 pKa = 10.7IEE29 pKa = 4.74NIDD32 pKa = 3.68YY33 pKa = 10.93KK34 pKa = 10.88DD35 pKa = 3.52VPRR38 pKa = 11.84LKK40 pKa = 10.42RR41 pKa = 11.84YY42 pKa = 10.01LSDD45 pKa = 3.23RR46 pKa = 11.84AKK48 pKa = 10.33IIPRR52 pKa = 11.84RR53 pKa = 11.84VTGTCARR60 pKa = 11.84HH61 pKa = 4.44QRR63 pKa = 11.84QLTVAVKK70 pKa = 10.26RR71 pKa = 11.84ARR73 pKa = 11.84HH74 pKa = 4.88VAFLPYY80 pKa = 10.57VGDD83 pKa = 3.67
MM1 pKa = 7.34EE2 pKa = 5.09KK3 pKa = 10.53EE4 pKa = 3.55RR5 pKa = 11.84TEE7 pKa = 4.12RR8 pKa = 11.84NDD10 pKa = 2.85RR11 pKa = 11.84RR12 pKa = 11.84NRR14 pKa = 11.84KK15 pKa = 8.07SRR17 pKa = 11.84KK18 pKa = 8.8KK19 pKa = 10.01VCAFCMDD26 pKa = 4.14KK27 pKa = 10.7IEE29 pKa = 4.74NIDD32 pKa = 3.68YY33 pKa = 10.93KK34 pKa = 10.88DD35 pKa = 3.52VPRR38 pKa = 11.84LKK40 pKa = 10.42RR41 pKa = 11.84YY42 pKa = 10.01LSDD45 pKa = 3.23RR46 pKa = 11.84AKK48 pKa = 10.33IIPRR52 pKa = 11.84RR53 pKa = 11.84VTGTCARR60 pKa = 11.84HH61 pKa = 4.44QRR63 pKa = 11.84QLTVAVKK70 pKa = 10.26RR71 pKa = 11.84ARR73 pKa = 11.84HH74 pKa = 4.88VAFLPYY80 pKa = 10.57VGDD83 pKa = 3.67
Molecular weight: 9.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
709936 |
30 |
1967 |
291.8 |
32.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.526 ± 0.054 | 1.762 ± 0.023 |
5.411 ± 0.036 | 7.26 ± 0.057 |
4.296 ± 0.04 | 6.89 ± 0.04 |
1.838 ± 0.024 | 6.967 ± 0.046 |
6.097 ± 0.043 | 9.556 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.051 ± 0.028 | 4.05 ± 0.033 |
3.648 ± 0.026 | 4.158 ± 0.037 |
4.313 ± 0.041 | 6.358 ± 0.05 |
5.356 ± 0.048 | 6.884 ± 0.036 |
0.907 ± 0.016 | 3.665 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
