
Alfalfa dwarf virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Alfalfa dwarf cytorhabdovirus
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
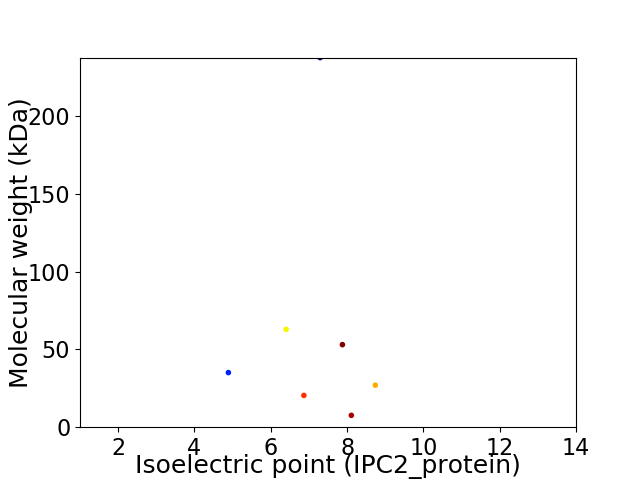
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F6QEW5|A0A0F6QEW5_9RHAB GDP polyribonucleotidyltransferase OS=Alfalfa dwarf virus OX=998864 PE=4 SV=1
MM1 pKa = 7.7AAPNFEE7 pKa = 4.75DD8 pKa = 4.15LSNPILDD15 pKa = 3.65MAMSGLNNSGDD26 pKa = 4.05EE27 pKa = 4.38DD28 pKa = 4.16SQIDD32 pKa = 3.64PAEE35 pKa = 4.34INDD38 pKa = 3.91DD39 pKa = 3.59VEE41 pKa = 4.52MGVEE45 pKa = 4.11NPKK48 pKa = 9.54MDD50 pKa = 3.63EE51 pKa = 4.18VAATDD56 pKa = 3.47AKK58 pKa = 10.98QPVDD62 pKa = 3.67EE63 pKa = 5.32YY64 pKa = 11.17EE65 pKa = 4.58IEE67 pKa = 5.47DD68 pKa = 4.09DD69 pKa = 4.87LAYY72 pKa = 10.78DD73 pKa = 3.93PADD76 pKa = 3.32VSHH79 pKa = 6.91ALEE82 pKa = 4.56DD83 pKa = 4.01LKK85 pKa = 11.16HH86 pKa = 6.04LCDD89 pKa = 3.73VRR91 pKa = 11.84GVNYY95 pKa = 7.52TVPMEE100 pKa = 4.16NQVSKK105 pKa = 11.01LFKK108 pKa = 10.93EE109 pKa = 4.61EE110 pKa = 3.84ILCYY114 pKa = 10.47SHH116 pKa = 6.87LKK118 pKa = 9.16WYY120 pKa = 9.89IRR122 pKa = 11.84GIILANQTQVLPTVTGAIADD142 pKa = 3.86MKK144 pKa = 10.6MEE146 pKa = 4.22TQHH149 pKa = 6.19LQKK152 pKa = 10.71SSNKK156 pKa = 8.73INKK159 pKa = 8.41EE160 pKa = 3.52VSRR163 pKa = 11.84PEE165 pKa = 3.67KK166 pKa = 9.39LTRR169 pKa = 11.84EE170 pKa = 3.97ITSEE174 pKa = 3.87LRR176 pKa = 11.84SIKK179 pKa = 10.58ADD181 pKa = 3.03IQEE184 pKa = 4.19SFRR187 pKa = 11.84ASMKK191 pKa = 10.7LFMDD195 pKa = 3.92EE196 pKa = 4.29TTTNSQRR203 pKa = 11.84LQPPVKK209 pKa = 9.94EE210 pKa = 4.24EE211 pKa = 3.96KK212 pKa = 9.63VTTNLISALVKK223 pKa = 10.6SKK225 pKa = 10.97EE226 pKa = 3.87KK227 pKa = 11.22AEE229 pKa = 4.33DD230 pKa = 3.67YY231 pKa = 10.75KK232 pKa = 11.62DD233 pKa = 3.37EE234 pKa = 4.32ATVSGVKK241 pKa = 8.89TAINIPKK248 pKa = 9.72PEE250 pKa = 4.17PSKK253 pKa = 10.71KK254 pKa = 10.56GRR256 pKa = 11.84DD257 pKa = 3.3GYY259 pKa = 9.64MAEE262 pKa = 4.69KK263 pKa = 8.56RR264 pKa = 11.84AHH266 pKa = 6.07MIRR269 pKa = 11.84YY270 pKa = 8.82GLDD273 pKa = 2.85ARR275 pKa = 11.84FVKK278 pKa = 9.62EE279 pKa = 4.17QPDD282 pKa = 3.86SVIDD286 pKa = 4.4LMYY289 pKa = 10.56PDD291 pKa = 5.31DD292 pKa = 4.01AHH294 pKa = 7.56SKK296 pKa = 9.34LKK298 pKa = 10.27QMKK301 pKa = 10.08LNNQQKK307 pKa = 9.45VEE309 pKa = 4.43GYY311 pKa = 10.3
MM1 pKa = 7.7AAPNFEE7 pKa = 4.75DD8 pKa = 4.15LSNPILDD15 pKa = 3.65MAMSGLNNSGDD26 pKa = 4.05EE27 pKa = 4.38DD28 pKa = 4.16SQIDD32 pKa = 3.64PAEE35 pKa = 4.34INDD38 pKa = 3.91DD39 pKa = 3.59VEE41 pKa = 4.52MGVEE45 pKa = 4.11NPKK48 pKa = 9.54MDD50 pKa = 3.63EE51 pKa = 4.18VAATDD56 pKa = 3.47AKK58 pKa = 10.98QPVDD62 pKa = 3.67EE63 pKa = 5.32YY64 pKa = 11.17EE65 pKa = 4.58IEE67 pKa = 5.47DD68 pKa = 4.09DD69 pKa = 4.87LAYY72 pKa = 10.78DD73 pKa = 3.93PADD76 pKa = 3.32VSHH79 pKa = 6.91ALEE82 pKa = 4.56DD83 pKa = 4.01LKK85 pKa = 11.16HH86 pKa = 6.04LCDD89 pKa = 3.73VRR91 pKa = 11.84GVNYY95 pKa = 7.52TVPMEE100 pKa = 4.16NQVSKK105 pKa = 11.01LFKK108 pKa = 10.93EE109 pKa = 4.61EE110 pKa = 3.84ILCYY114 pKa = 10.47SHH116 pKa = 6.87LKK118 pKa = 9.16WYY120 pKa = 9.89IRR122 pKa = 11.84GIILANQTQVLPTVTGAIADD142 pKa = 3.86MKK144 pKa = 10.6MEE146 pKa = 4.22TQHH149 pKa = 6.19LQKK152 pKa = 10.71SSNKK156 pKa = 8.73INKK159 pKa = 8.41EE160 pKa = 3.52VSRR163 pKa = 11.84PEE165 pKa = 3.67KK166 pKa = 9.39LTRR169 pKa = 11.84EE170 pKa = 3.97ITSEE174 pKa = 3.87LRR176 pKa = 11.84SIKK179 pKa = 10.58ADD181 pKa = 3.03IQEE184 pKa = 4.19SFRR187 pKa = 11.84ASMKK191 pKa = 10.7LFMDD195 pKa = 3.92EE196 pKa = 4.29TTTNSQRR203 pKa = 11.84LQPPVKK209 pKa = 9.94EE210 pKa = 4.24EE211 pKa = 3.96KK212 pKa = 9.63VTTNLISALVKK223 pKa = 10.6SKK225 pKa = 10.97EE226 pKa = 3.87KK227 pKa = 11.22AEE229 pKa = 4.33DD230 pKa = 3.67YY231 pKa = 10.75KK232 pKa = 11.62DD233 pKa = 3.37EE234 pKa = 4.32ATVSGVKK241 pKa = 8.89TAINIPKK248 pKa = 9.72PEE250 pKa = 4.17PSKK253 pKa = 10.71KK254 pKa = 10.56GRR256 pKa = 11.84DD257 pKa = 3.3GYY259 pKa = 9.64MAEE262 pKa = 4.69KK263 pKa = 8.56RR264 pKa = 11.84AHH266 pKa = 6.07MIRR269 pKa = 11.84YY270 pKa = 8.82GLDD273 pKa = 2.85ARR275 pKa = 11.84FVKK278 pKa = 9.62EE279 pKa = 4.17QPDD282 pKa = 3.86SVIDD286 pKa = 4.4LMYY289 pKa = 10.56PDD291 pKa = 5.31DD292 pKa = 4.01AHH294 pKa = 7.56SKK296 pKa = 9.34LKK298 pKa = 10.27QMKK301 pKa = 10.08LNNQQKK307 pKa = 9.45VEE309 pKa = 4.43GYY311 pKa = 10.3
Molecular weight: 35.13 kDa
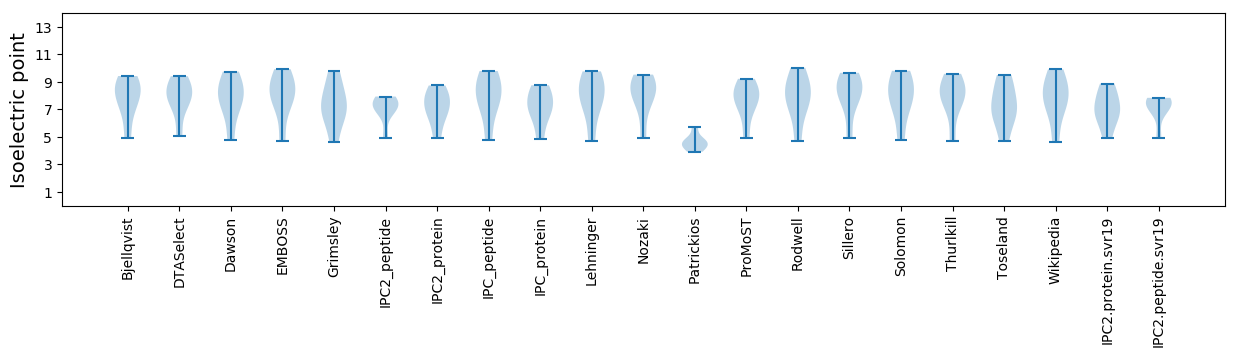
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F6QEW1|A0A0F6QEW1_9RHAB Phosphoprotein OS=Alfalfa dwarf virus OX=998864 PE=4 SV=1
MM1 pKa = 7.98PIRR4 pKa = 11.84VGSTSYY10 pKa = 10.19TLDD13 pKa = 3.32KK14 pKa = 10.5EE15 pKa = 4.47LVVGAEE21 pKa = 3.97NGEE24 pKa = 4.24VPLTKK29 pKa = 9.94TLSLFQRR36 pKa = 11.84ATMLNAKK43 pKa = 9.7NYY45 pKa = 9.96RR46 pKa = 11.84IKK48 pKa = 10.47QLRR51 pKa = 11.84FVYY54 pKa = 10.19KK55 pKa = 10.16PRR57 pKa = 11.84TGSLGEE63 pKa = 4.03GTIEE67 pKa = 4.65TRR69 pKa = 11.84VKK71 pKa = 10.8DD72 pKa = 3.48NRR74 pKa = 11.84IDD76 pKa = 3.64SRR78 pKa = 11.84ISDD81 pKa = 4.42PIINRR86 pKa = 11.84FRR88 pKa = 11.84FDD90 pKa = 3.23ASASADD96 pKa = 3.98LKK98 pKa = 10.33WEE100 pKa = 4.03SSFWIPKK107 pKa = 10.14DD108 pKa = 3.57DD109 pKa = 4.17FKK111 pKa = 11.63SGTKK115 pKa = 10.34APIILEE121 pKa = 4.13TDD123 pKa = 3.39VLEE126 pKa = 4.95CNLQEE131 pKa = 4.78GYY133 pKa = 10.68SIGLLSIIVSVSASDD148 pKa = 3.29TMDD151 pKa = 2.94NFIYY155 pKa = 10.23RR156 pKa = 11.84RR157 pKa = 11.84PSAEE161 pKa = 3.44IRR163 pKa = 11.84EE164 pKa = 4.35DD165 pKa = 3.33PFKK168 pKa = 10.81ILAVGGSKK176 pKa = 10.07SAQSKK181 pKa = 9.27FIQTPVQRR189 pKa = 11.84MITDD193 pKa = 3.67EE194 pKa = 4.07QTGAPEE200 pKa = 3.9DD201 pKa = 4.41AEE203 pKa = 4.55MKK205 pKa = 10.0NHH207 pKa = 6.03LRR209 pKa = 11.84VIARR213 pKa = 11.84DD214 pKa = 3.81SVGSRR219 pKa = 11.84SSRR222 pKa = 11.84MIEE225 pKa = 3.65PARR228 pKa = 11.84PSGYY232 pKa = 7.45RR233 pKa = 11.84QRR235 pKa = 11.84RR236 pKa = 11.84IRR238 pKa = 11.84EE239 pKa = 3.93VV240 pKa = 2.91
MM1 pKa = 7.98PIRR4 pKa = 11.84VGSTSYY10 pKa = 10.19TLDD13 pKa = 3.32KK14 pKa = 10.5EE15 pKa = 4.47LVVGAEE21 pKa = 3.97NGEE24 pKa = 4.24VPLTKK29 pKa = 9.94TLSLFQRR36 pKa = 11.84ATMLNAKK43 pKa = 9.7NYY45 pKa = 9.96RR46 pKa = 11.84IKK48 pKa = 10.47QLRR51 pKa = 11.84FVYY54 pKa = 10.19KK55 pKa = 10.16PRR57 pKa = 11.84TGSLGEE63 pKa = 4.03GTIEE67 pKa = 4.65TRR69 pKa = 11.84VKK71 pKa = 10.8DD72 pKa = 3.48NRR74 pKa = 11.84IDD76 pKa = 3.64SRR78 pKa = 11.84ISDD81 pKa = 4.42PIINRR86 pKa = 11.84FRR88 pKa = 11.84FDD90 pKa = 3.23ASASADD96 pKa = 3.98LKK98 pKa = 10.33WEE100 pKa = 4.03SSFWIPKK107 pKa = 10.14DD108 pKa = 3.57DD109 pKa = 4.17FKK111 pKa = 11.63SGTKK115 pKa = 10.34APIILEE121 pKa = 4.13TDD123 pKa = 3.39VLEE126 pKa = 4.95CNLQEE131 pKa = 4.78GYY133 pKa = 10.68SIGLLSIIVSVSASDD148 pKa = 3.29TMDD151 pKa = 2.94NFIYY155 pKa = 10.23RR156 pKa = 11.84RR157 pKa = 11.84PSAEE161 pKa = 3.44IRR163 pKa = 11.84EE164 pKa = 4.35DD165 pKa = 3.33PFKK168 pKa = 10.81ILAVGGSKK176 pKa = 10.07SAQSKK181 pKa = 9.27FIQTPVQRR189 pKa = 11.84MITDD193 pKa = 3.67EE194 pKa = 4.07QTGAPEE200 pKa = 3.9DD201 pKa = 4.41AEE203 pKa = 4.55MKK205 pKa = 10.0NHH207 pKa = 6.03LRR209 pKa = 11.84VIARR213 pKa = 11.84DD214 pKa = 3.81SVGSRR219 pKa = 11.84SSRR222 pKa = 11.84MIEE225 pKa = 3.65PARR228 pKa = 11.84PSGYY232 pKa = 7.45RR233 pKa = 11.84QRR235 pKa = 11.84RR236 pKa = 11.84IRR238 pKa = 11.84EE239 pKa = 3.93VV240 pKa = 2.91
Molecular weight: 27.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3937 |
65 |
2085 |
562.4 |
63.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.918 ± 0.94 | 1.88 ± 0.332 |
6.655 ± 0.383 | 5.867 ± 0.435 |
2.87 ± 0.246 | 5.994 ± 0.357 |
2.032 ± 0.276 | 6.528 ± 0.396 |
6.883 ± 0.413 | 9.093 ± 0.588 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.607 ± 0.404 | 3.759 ± 0.277 |
4.496 ± 0.294 | 2.769 ± 0.278 |
5.817 ± 0.496 | 7.823 ± 0.519 |
6.452 ± 0.369 | 6.452 ± 0.217 |
1.575 ± 0.242 | 3.531 ± 0.259 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
