
Pomona bat hepatitis B virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; Orthohepadnavirus
Average proteome isoelectric point is 8.82
Get precalculated fractions of proteins
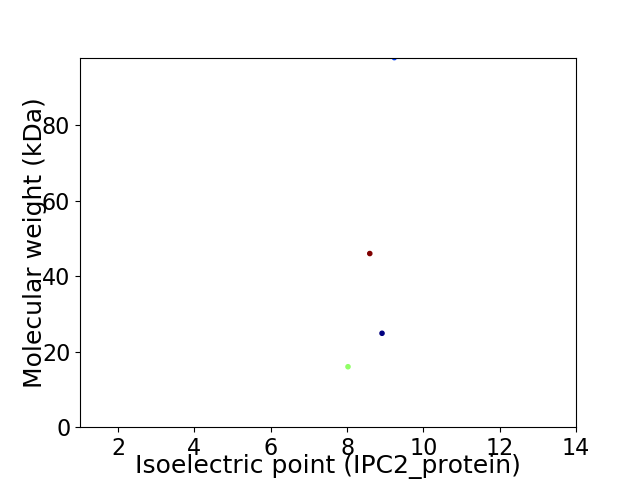
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7DQ32|A0A0A7DQ32_9HEPA DNA-directed DNA polymerase OS=Pomona bat hepatitis B virus OX=2049933 PE=3 SV=1
MM1 pKa = 7.76AARR4 pKa = 11.84LYY6 pKa = 11.12CEE8 pKa = 5.34LDD10 pKa = 3.32PSRR13 pKa = 11.84DD14 pKa = 3.65VLLLRR19 pKa = 11.84PFGAQPGGRR28 pKa = 11.84PVSRR32 pKa = 11.84FPGDD36 pKa = 3.76SNGSTASTVPPVHH49 pKa = 6.39RR50 pKa = 11.84QDD52 pKa = 3.1FALRR56 pKa = 11.84RR57 pKa = 11.84LPACAYY63 pKa = 10.34SNAGPCVLRR72 pKa = 11.84FTCAEE77 pKa = 3.74AAVNMEE83 pKa = 3.9TTMTNFVTWHH93 pKa = 5.34TARR96 pKa = 11.84QRR98 pKa = 11.84GTVTQTLNSWQWYY111 pKa = 8.95FGQLLMNQWEE121 pKa = 4.34EE122 pKa = 4.25TGWDD126 pKa = 3.64DD127 pKa = 5.84RR128 pKa = 11.84IIAYY132 pKa = 10.07VLGGCRR138 pKa = 11.84HH139 pKa = 6.37KK140 pKa = 11.13LRR142 pKa = 5.47
MM1 pKa = 7.76AARR4 pKa = 11.84LYY6 pKa = 11.12CEE8 pKa = 5.34LDD10 pKa = 3.32PSRR13 pKa = 11.84DD14 pKa = 3.65VLLLRR19 pKa = 11.84PFGAQPGGRR28 pKa = 11.84PVSRR32 pKa = 11.84FPGDD36 pKa = 3.76SNGSTASTVPPVHH49 pKa = 6.39RR50 pKa = 11.84QDD52 pKa = 3.1FALRR56 pKa = 11.84RR57 pKa = 11.84LPACAYY63 pKa = 10.34SNAGPCVLRR72 pKa = 11.84FTCAEE77 pKa = 3.74AAVNMEE83 pKa = 3.9TTMTNFVTWHH93 pKa = 5.34TARR96 pKa = 11.84QRR98 pKa = 11.84GTVTQTLNSWQWYY111 pKa = 8.95FGQLLMNQWEE121 pKa = 4.34EE122 pKa = 4.25TGWDD126 pKa = 3.64DD127 pKa = 5.84RR128 pKa = 11.84IIAYY132 pKa = 10.07VLGGCRR138 pKa = 11.84HH139 pKa = 6.37KK140 pKa = 11.13LRR142 pKa = 5.47
Molecular weight: 16.01 kDa
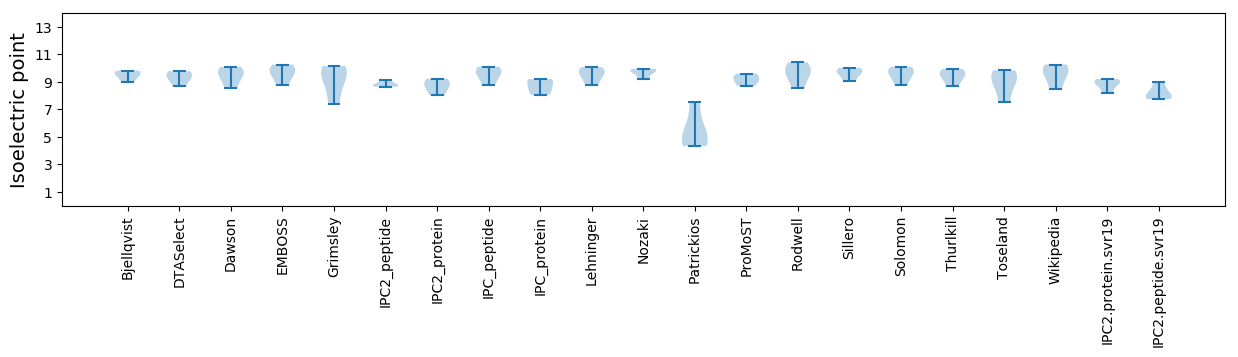
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7DQC0|A0A0A7DQC0_9HEPA External core antigen OS=Pomona bat hepatitis B virus OX=2049933 GN=C PE=3 SV=1
MM1 pKa = 7.55HH2 pKa = 7.82PFSQLFQSIQSLEE15 pKa = 4.11EE16 pKa = 4.42GEE18 pKa = 4.59VEE20 pKa = 4.27QLLGPPEE27 pKa = 4.38DD28 pKa = 4.57ALPLLADD35 pKa = 3.84ANLNHH40 pKa = 6.66RR41 pKa = 11.84VAEE44 pKa = 4.07EE45 pKa = 4.32LLLQLPLHH53 pKa = 6.24AEE55 pKa = 3.76IWVHH59 pKa = 6.08KK60 pKa = 10.62AGGLTGLYY68 pKa = 9.74SEE70 pKa = 5.12KK71 pKa = 10.54PKK73 pKa = 10.56IFNPEE78 pKa = 3.15WKK80 pKa = 8.77TPSFPNIHH88 pKa = 6.47LRR90 pKa = 11.84HH91 pKa = 6.63DD92 pKa = 3.78LCEE95 pKa = 5.37HH96 pKa = 5.87IDD98 pKa = 3.61SRR100 pKa = 11.84IGLTKK105 pKa = 10.22GEE107 pKa = 3.86KK108 pKa = 9.78RR109 pKa = 11.84RR110 pKa = 11.84FKK112 pKa = 10.82LCLPARR118 pKa = 11.84FFPNKK123 pKa = 8.53TKK125 pKa = 10.77YY126 pKa = 10.22FPLHH130 pKa = 5.72NAIKK134 pKa = 10.3NYY136 pKa = 10.35YY137 pKa = 8.12PDD139 pKa = 3.18HH140 pKa = 6.37TLTHH144 pKa = 6.93FFHH147 pKa = 6.52TAHH150 pKa = 6.33YY151 pKa = 10.36LWTLWEE157 pKa = 4.25SGVTYY162 pKa = 10.69LRR164 pKa = 11.84TSQSSASFNGQSYY177 pKa = 8.76PWEE180 pKa = 4.46QKK182 pKa = 9.78HH183 pKa = 5.4HH184 pKa = 4.65THH186 pKa = 6.75GKK188 pKa = 7.25QQISSQSLRR197 pKa = 11.84LPPRR201 pKa = 11.84SPATNSPKK209 pKa = 9.96FRR211 pKa = 11.84GLGKK215 pKa = 10.18ILGPKK220 pKa = 8.23SAKK223 pKa = 10.15GSLAPSIGTLSGSLGTKK240 pKa = 10.07SSPSSWGSTWNRR252 pKa = 11.84RR253 pKa = 11.84EE254 pKa = 4.09FSGSPYY260 pKa = 10.11QYY262 pKa = 10.6QKK264 pKa = 11.06YY265 pKa = 9.48RR266 pKa = 11.84RR267 pKa = 11.84NNQNYY272 pKa = 8.58YY273 pKa = 9.26RR274 pKa = 11.84QKK276 pKa = 7.76TTNNYY281 pKa = 9.36YY282 pKa = 10.58KK283 pKa = 10.61NKK285 pKa = 9.23QDD287 pKa = 4.88QSIYY291 pKa = 9.64WNNANKK297 pKa = 9.9TNITSHH303 pKa = 6.27TSIQSPKK310 pKa = 10.06GASTNAPDD318 pKa = 3.62ITSKK322 pKa = 9.96TDD324 pKa = 2.9SSTSQYY330 pKa = 10.62EE331 pKa = 3.76QDD333 pKa = 3.91LEE335 pKa = 4.63TEE337 pKa = 4.3VRR339 pKa = 11.84QPDD342 pKa = 3.96NSCPWLFFNHH352 pKa = 5.97SPACGRR358 pKa = 11.84YY359 pKa = 9.31CLRR362 pKa = 11.84QCLLLLSDD370 pKa = 4.24WGPCHH375 pKa = 6.75RR376 pKa = 11.84HH377 pKa = 5.42GDD379 pKa = 3.72HH380 pKa = 8.01LIRR383 pKa = 11.84TPRR386 pKa = 11.84TPRR389 pKa = 11.84RR390 pKa = 11.84ISGGVFLVDD399 pKa = 3.54KK400 pKa = 10.57NPNNRR405 pKa = 11.84TEE407 pKa = 3.84SRR409 pKa = 11.84LVVDD413 pKa = 5.53FSQFSRR419 pKa = 11.84GNTRR423 pKa = 11.84VQWPKK428 pKa = 10.19FAVPNLQALTNLLSSNLSWLSLDD451 pKa = 3.44VSAAFYY457 pKa = 9.93HH458 pKa = 6.73IPLSPAAMPHH468 pKa = 6.39LLVGSPGLEE477 pKa = 3.7GLASCMSPKK486 pKa = 9.38STNNNHH492 pKa = 6.07QEE494 pKa = 4.08MQHH497 pKa = 5.46LHH499 pKa = 5.34SLCGRR504 pKa = 11.84RR505 pKa = 11.84NLLGLLLLFKK515 pKa = 9.84TCRR518 pKa = 11.84RR519 pKa = 11.84KK520 pKa = 8.58QTLLGHH526 pKa = 5.9SFFKK530 pKa = 10.76GFRR533 pKa = 11.84KK534 pKa = 9.91IPTGVGLSPILLAPFPSALASVVRR558 pKa = 11.84RR559 pKa = 11.84TFPHH563 pKa = 6.58CLAFTYY569 pKa = 10.09MDD571 pKa = 4.38DD572 pKa = 3.8VVLGATTPPHH582 pKa = 6.63LEE584 pKa = 3.64SVYY587 pKa = 11.26ASVCSLFSDD596 pKa = 4.42LGIVLNPDD604 pKa = 2.8KK605 pKa = 10.83TKK607 pKa = 9.36WWGKK611 pKa = 8.82HH612 pKa = 4.06LQFMGLLITPSGTLPQSKK630 pKa = 7.16QQKK633 pKa = 8.22WAIQLLKK640 pKa = 10.68ALPCYY645 pKa = 10.25TIFDD649 pKa = 3.79WKK651 pKa = 10.31ILQRR655 pKa = 11.84LTGLLGFLAPFTACGYY671 pKa = 7.21PALMPLYY678 pKa = 10.4HH679 pKa = 7.76AITQKK684 pKa = 10.97NGFEE688 pKa = 4.06FSPAYY693 pKa = 9.78KK694 pKa = 10.59GFLLRR699 pKa = 11.84LYY701 pKa = 10.38SRR703 pKa = 11.84PLHH706 pKa = 5.4VVRR709 pKa = 11.84QRR711 pKa = 11.84SSTCQVFADD720 pKa = 4.26ATPTGWGLVNHH731 pKa = 5.94QLALYY736 pKa = 10.16KK737 pKa = 10.21SGRR740 pKa = 11.84FPRR743 pKa = 11.84PLPIHH748 pKa = 5.89HH749 pKa = 7.41AEE751 pKa = 4.71LIAAILARR759 pKa = 11.84CWSGAKK765 pKa = 10.49LLGTDD770 pKa = 3.34NTIVCSHH777 pKa = 6.99KK778 pKa = 9.65FTHH781 pKa = 6.59FPWLLGCTANWILRR795 pKa = 11.84EE796 pKa = 3.91TSFCYY801 pKa = 10.56VPSEE805 pKa = 4.58LNPADD810 pKa = 3.74APSRR814 pKa = 11.84GFLGIRR820 pKa = 11.84TAPPPLRR827 pKa = 11.84FRR829 pKa = 11.84PSTGRR834 pKa = 11.84TSLFVVSPPAPTRR847 pKa = 11.84MPDD850 pKa = 2.88RR851 pKa = 11.84VSFVSPVQKK860 pKa = 10.5RR861 pKa = 11.84LSTWRR866 pKa = 11.84PPP868 pKa = 3.22
MM1 pKa = 7.55HH2 pKa = 7.82PFSQLFQSIQSLEE15 pKa = 4.11EE16 pKa = 4.42GEE18 pKa = 4.59VEE20 pKa = 4.27QLLGPPEE27 pKa = 4.38DD28 pKa = 4.57ALPLLADD35 pKa = 3.84ANLNHH40 pKa = 6.66RR41 pKa = 11.84VAEE44 pKa = 4.07EE45 pKa = 4.32LLLQLPLHH53 pKa = 6.24AEE55 pKa = 3.76IWVHH59 pKa = 6.08KK60 pKa = 10.62AGGLTGLYY68 pKa = 9.74SEE70 pKa = 5.12KK71 pKa = 10.54PKK73 pKa = 10.56IFNPEE78 pKa = 3.15WKK80 pKa = 8.77TPSFPNIHH88 pKa = 6.47LRR90 pKa = 11.84HH91 pKa = 6.63DD92 pKa = 3.78LCEE95 pKa = 5.37HH96 pKa = 5.87IDD98 pKa = 3.61SRR100 pKa = 11.84IGLTKK105 pKa = 10.22GEE107 pKa = 3.86KK108 pKa = 9.78RR109 pKa = 11.84RR110 pKa = 11.84FKK112 pKa = 10.82LCLPARR118 pKa = 11.84FFPNKK123 pKa = 8.53TKK125 pKa = 10.77YY126 pKa = 10.22FPLHH130 pKa = 5.72NAIKK134 pKa = 10.3NYY136 pKa = 10.35YY137 pKa = 8.12PDD139 pKa = 3.18HH140 pKa = 6.37TLTHH144 pKa = 6.93FFHH147 pKa = 6.52TAHH150 pKa = 6.33YY151 pKa = 10.36LWTLWEE157 pKa = 4.25SGVTYY162 pKa = 10.69LRR164 pKa = 11.84TSQSSASFNGQSYY177 pKa = 8.76PWEE180 pKa = 4.46QKK182 pKa = 9.78HH183 pKa = 5.4HH184 pKa = 4.65THH186 pKa = 6.75GKK188 pKa = 7.25QQISSQSLRR197 pKa = 11.84LPPRR201 pKa = 11.84SPATNSPKK209 pKa = 9.96FRR211 pKa = 11.84GLGKK215 pKa = 10.18ILGPKK220 pKa = 8.23SAKK223 pKa = 10.15GSLAPSIGTLSGSLGTKK240 pKa = 10.07SSPSSWGSTWNRR252 pKa = 11.84RR253 pKa = 11.84EE254 pKa = 4.09FSGSPYY260 pKa = 10.11QYY262 pKa = 10.6QKK264 pKa = 11.06YY265 pKa = 9.48RR266 pKa = 11.84RR267 pKa = 11.84NNQNYY272 pKa = 8.58YY273 pKa = 9.26RR274 pKa = 11.84QKK276 pKa = 7.76TTNNYY281 pKa = 9.36YY282 pKa = 10.58KK283 pKa = 10.61NKK285 pKa = 9.23QDD287 pKa = 4.88QSIYY291 pKa = 9.64WNNANKK297 pKa = 9.9TNITSHH303 pKa = 6.27TSIQSPKK310 pKa = 10.06GASTNAPDD318 pKa = 3.62ITSKK322 pKa = 9.96TDD324 pKa = 2.9SSTSQYY330 pKa = 10.62EE331 pKa = 3.76QDD333 pKa = 3.91LEE335 pKa = 4.63TEE337 pKa = 4.3VRR339 pKa = 11.84QPDD342 pKa = 3.96NSCPWLFFNHH352 pKa = 5.97SPACGRR358 pKa = 11.84YY359 pKa = 9.31CLRR362 pKa = 11.84QCLLLLSDD370 pKa = 4.24WGPCHH375 pKa = 6.75RR376 pKa = 11.84HH377 pKa = 5.42GDD379 pKa = 3.72HH380 pKa = 8.01LIRR383 pKa = 11.84TPRR386 pKa = 11.84TPRR389 pKa = 11.84RR390 pKa = 11.84ISGGVFLVDD399 pKa = 3.54KK400 pKa = 10.57NPNNRR405 pKa = 11.84TEE407 pKa = 3.84SRR409 pKa = 11.84LVVDD413 pKa = 5.53FSQFSRR419 pKa = 11.84GNTRR423 pKa = 11.84VQWPKK428 pKa = 10.19FAVPNLQALTNLLSSNLSWLSLDD451 pKa = 3.44VSAAFYY457 pKa = 9.93HH458 pKa = 6.73IPLSPAAMPHH468 pKa = 6.39LLVGSPGLEE477 pKa = 3.7GLASCMSPKK486 pKa = 9.38STNNNHH492 pKa = 6.07QEE494 pKa = 4.08MQHH497 pKa = 5.46LHH499 pKa = 5.34SLCGRR504 pKa = 11.84RR505 pKa = 11.84NLLGLLLLFKK515 pKa = 9.84TCRR518 pKa = 11.84RR519 pKa = 11.84KK520 pKa = 8.58QTLLGHH526 pKa = 5.9SFFKK530 pKa = 10.76GFRR533 pKa = 11.84KK534 pKa = 9.91IPTGVGLSPILLAPFPSALASVVRR558 pKa = 11.84RR559 pKa = 11.84TFPHH563 pKa = 6.58CLAFTYY569 pKa = 10.09MDD571 pKa = 4.38DD572 pKa = 3.8VVLGATTPPHH582 pKa = 6.63LEE584 pKa = 3.64SVYY587 pKa = 11.26ASVCSLFSDD596 pKa = 4.42LGIVLNPDD604 pKa = 2.8KK605 pKa = 10.83TKK607 pKa = 9.36WWGKK611 pKa = 8.82HH612 pKa = 4.06LQFMGLLITPSGTLPQSKK630 pKa = 7.16QQKK633 pKa = 8.22WAIQLLKK640 pKa = 10.68ALPCYY645 pKa = 10.25TIFDD649 pKa = 3.79WKK651 pKa = 10.31ILQRR655 pKa = 11.84LTGLLGFLAPFTACGYY671 pKa = 7.21PALMPLYY678 pKa = 10.4HH679 pKa = 7.76AITQKK684 pKa = 10.97NGFEE688 pKa = 4.06FSPAYY693 pKa = 9.78KK694 pKa = 10.59GFLLRR699 pKa = 11.84LYY701 pKa = 10.38SRR703 pKa = 11.84PLHH706 pKa = 5.4VVRR709 pKa = 11.84QRR711 pKa = 11.84SSTCQVFADD720 pKa = 4.26ATPTGWGLVNHH731 pKa = 5.94QLALYY736 pKa = 10.16KK737 pKa = 10.21SGRR740 pKa = 11.84FPRR743 pKa = 11.84PLPIHH748 pKa = 5.89HH749 pKa = 7.41AEE751 pKa = 4.71LIAAILARR759 pKa = 11.84CWSGAKK765 pKa = 10.49LLGTDD770 pKa = 3.34NTIVCSHH777 pKa = 6.99KK778 pKa = 9.65FTHH781 pKa = 6.59FPWLLGCTANWILRR795 pKa = 11.84EE796 pKa = 3.91TSFCYY801 pKa = 10.56VPSEE805 pKa = 4.58LNPADD810 pKa = 3.74APSRR814 pKa = 11.84GFLGIRR820 pKa = 11.84TAPPPLRR827 pKa = 11.84FRR829 pKa = 11.84PSTGRR834 pKa = 11.84TSLFVVSPPAPTRR847 pKa = 11.84MPDD850 pKa = 2.88RR851 pKa = 11.84VSFVSPVQKK860 pKa = 10.5RR861 pKa = 11.84LSTWRR866 pKa = 11.84PPP868 pKa = 3.22
Molecular weight: 97.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1642 |
142 |
868 |
410.5 |
46.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.725 ± 0.382 | 2.741 ± 0.338 |
2.862 ± 0.147 | 2.984 ± 0.392 |
4.75 ± 0.279 | 6.395 ± 0.344 |
3.228 ± 0.58 | 3.837 ± 0.39 |
4.019 ± 0.748 | 12.119 ± 0.488 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.34 ± 0.264 | 4.141 ± 0.402 |
8.709 ± 0.431 | 4.202 ± 0.233 |
6.456 ± 0.917 | 8.952 ± 0.493 |
7.734 ± 0.655 | 4.141 ± 0.47 |
3.167 ± 0.526 | 2.497 ± 0.426 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
