
Cryptosporidium parvum virus 1 (strain KSU-1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Cryspovirus; Cryptosporidium parvum virus 1
Average proteome isoelectric point is 7.9
Get precalculated fractions of proteins
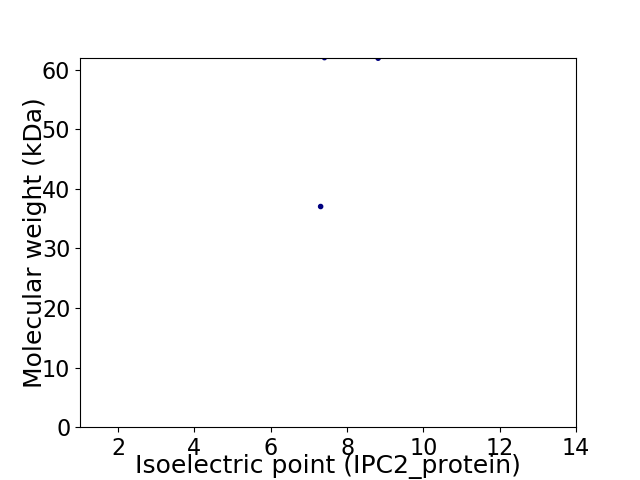
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
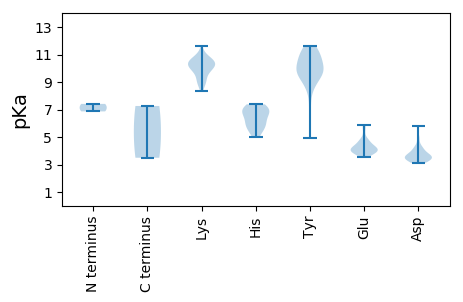
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|O15926|CAPSD_CPVKS Capsid protein OS=Cryptosporidium parvum virus 1 (strain KSU-1) OX=766191 PE=3 SV=1
MM1 pKa = 6.86ITSFEE6 pKa = 4.33SIEE9 pKa = 4.24KK10 pKa = 10.25KK11 pKa = 10.13NAVYY15 pKa = 10.85YY16 pKa = 10.09PVDD19 pKa = 3.83LKK21 pKa = 11.19FVTDD25 pKa = 4.48LSSDD29 pKa = 3.75LSNTADD35 pKa = 3.45GLGQAWYY42 pKa = 9.68KK43 pKa = 9.96ISQVAVEE50 pKa = 4.83HH51 pKa = 6.98IILTALKK58 pKa = 9.58INYY61 pKa = 8.35VLNHH65 pKa = 5.94RR66 pKa = 11.84TLLLIYY72 pKa = 9.68NKK74 pKa = 9.91KK75 pKa = 9.59VPDD78 pKa = 3.55MDD80 pKa = 5.81LINIIQSSIMVPRR93 pKa = 11.84FVRR96 pKa = 11.84DD97 pKa = 4.0LIRR100 pKa = 11.84EE101 pKa = 4.07ILRR104 pKa = 11.84PMHH107 pKa = 6.58HH108 pKa = 6.69SGITYY113 pKa = 10.0IPDD116 pKa = 3.93LDD118 pKa = 4.3LSTRR122 pKa = 11.84PTPHH126 pKa = 7.38LLDD129 pKa = 3.31TFYY132 pKa = 10.66PIAEE136 pKa = 4.42HH137 pKa = 5.41LTRR140 pKa = 11.84WNNVCQKK147 pKa = 10.48LGFEE151 pKa = 4.64MVPILPEE158 pKa = 3.98AVQSVSLTFYY168 pKa = 10.79SYY170 pKa = 10.01EE171 pKa = 4.0TDD173 pKa = 3.19EE174 pKa = 5.57LLSFDD179 pKa = 4.89NLINLDD185 pKa = 3.26WRR187 pKa = 11.84IEE189 pKa = 3.93AFGWTKK195 pKa = 10.68HH196 pKa = 5.83LVHH199 pKa = 6.7NPTSEE204 pKa = 3.74VDD206 pKa = 3.65EE207 pKa = 4.81LGKK210 pKa = 10.41SQTATSRR217 pKa = 11.84KK218 pKa = 8.62RR219 pKa = 11.84VQEE222 pKa = 3.74KK223 pKa = 10.46DD224 pKa = 3.55YY225 pKa = 11.36EE226 pKa = 4.83CDD228 pKa = 3.38DD229 pKa = 3.47FRR231 pKa = 11.84KK232 pKa = 10.17VYY234 pKa = 9.74EE235 pKa = 4.31RR236 pKa = 11.84PLQNRR241 pKa = 11.84RR242 pKa = 11.84ILGMIVYY249 pKa = 9.77RR250 pKa = 11.84YY251 pKa = 7.17TCAPYY256 pKa = 10.54TMRR259 pKa = 11.84LGHH262 pKa = 7.15LSPNCRR268 pKa = 11.84FPTEE272 pKa = 4.38KK273 pKa = 10.25NHH275 pKa = 6.78SEE277 pKa = 4.15TPPTSNRR284 pKa = 11.84VLLSEE289 pKa = 3.85QTMVHH294 pKa = 6.68SIQKK298 pKa = 9.38NRR300 pKa = 11.84SKK302 pKa = 10.45PKK304 pKa = 8.99KK305 pKa = 8.57VKK307 pKa = 10.18IVTTEE312 pKa = 3.97CSADD316 pKa = 3.57RR317 pKa = 11.84SHH319 pKa = 7.26
MM1 pKa = 6.86ITSFEE6 pKa = 4.33SIEE9 pKa = 4.24KK10 pKa = 10.25KK11 pKa = 10.13NAVYY15 pKa = 10.85YY16 pKa = 10.09PVDD19 pKa = 3.83LKK21 pKa = 11.19FVTDD25 pKa = 4.48LSSDD29 pKa = 3.75LSNTADD35 pKa = 3.45GLGQAWYY42 pKa = 9.68KK43 pKa = 9.96ISQVAVEE50 pKa = 4.83HH51 pKa = 6.98IILTALKK58 pKa = 9.58INYY61 pKa = 8.35VLNHH65 pKa = 5.94RR66 pKa = 11.84TLLLIYY72 pKa = 9.68NKK74 pKa = 9.91KK75 pKa = 9.59VPDD78 pKa = 3.55MDD80 pKa = 5.81LINIIQSSIMVPRR93 pKa = 11.84FVRR96 pKa = 11.84DD97 pKa = 4.0LIRR100 pKa = 11.84EE101 pKa = 4.07ILRR104 pKa = 11.84PMHH107 pKa = 6.58HH108 pKa = 6.69SGITYY113 pKa = 10.0IPDD116 pKa = 3.93LDD118 pKa = 4.3LSTRR122 pKa = 11.84PTPHH126 pKa = 7.38LLDD129 pKa = 3.31TFYY132 pKa = 10.66PIAEE136 pKa = 4.42HH137 pKa = 5.41LTRR140 pKa = 11.84WNNVCQKK147 pKa = 10.48LGFEE151 pKa = 4.64MVPILPEE158 pKa = 3.98AVQSVSLTFYY168 pKa = 10.79SYY170 pKa = 10.01EE171 pKa = 4.0TDD173 pKa = 3.19EE174 pKa = 5.57LLSFDD179 pKa = 4.89NLINLDD185 pKa = 3.26WRR187 pKa = 11.84IEE189 pKa = 3.93AFGWTKK195 pKa = 10.68HH196 pKa = 5.83LVHH199 pKa = 6.7NPTSEE204 pKa = 3.74VDD206 pKa = 3.65EE207 pKa = 4.81LGKK210 pKa = 10.41SQTATSRR217 pKa = 11.84KK218 pKa = 8.62RR219 pKa = 11.84VQEE222 pKa = 3.74KK223 pKa = 10.46DD224 pKa = 3.55YY225 pKa = 11.36EE226 pKa = 4.83CDD228 pKa = 3.38DD229 pKa = 3.47FRR231 pKa = 11.84KK232 pKa = 10.17VYY234 pKa = 9.74EE235 pKa = 4.31RR236 pKa = 11.84PLQNRR241 pKa = 11.84RR242 pKa = 11.84ILGMIVYY249 pKa = 9.77RR250 pKa = 11.84YY251 pKa = 7.17TCAPYY256 pKa = 10.54TMRR259 pKa = 11.84LGHH262 pKa = 7.15LSPNCRR268 pKa = 11.84FPTEE272 pKa = 4.38KK273 pKa = 10.25NHH275 pKa = 6.78SEE277 pKa = 4.15TPPTSNRR284 pKa = 11.84VLLSEE289 pKa = 3.85QTMVHH294 pKa = 6.68SIQKK298 pKa = 9.38NRR300 pKa = 11.84SKK302 pKa = 10.45PKK304 pKa = 8.99KK305 pKa = 8.57VKK307 pKa = 10.18IVTTEE312 pKa = 3.97CSADD316 pKa = 3.57RR317 pKa = 11.84SHH319 pKa = 7.26
Molecular weight: 37.03 kDa
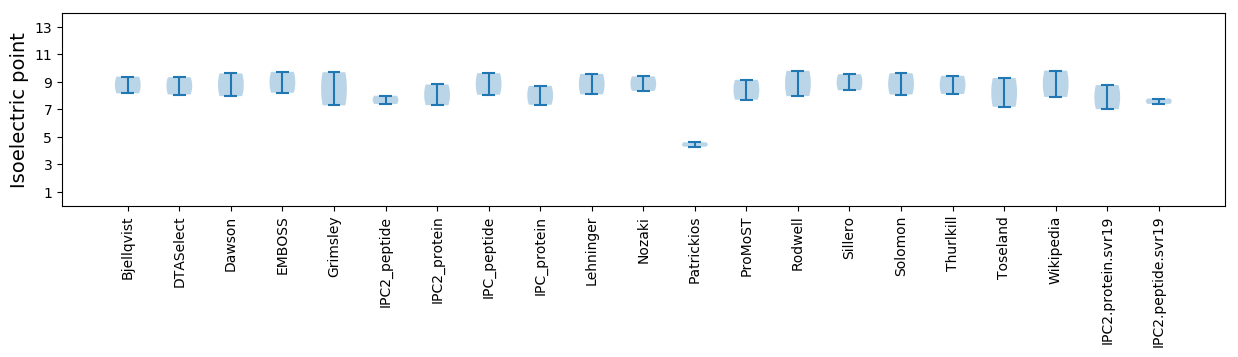
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O15926|CAPSD_CPVKS Capsid protein OS=Cryptosporidium parvum virus 1 (strain KSU-1) OX=766191 PE=3 SV=1
MM1 pKa = 7.41KK2 pKa = 10.09FVNIYY7 pKa = 9.6EE8 pKa = 4.09IQRR11 pKa = 11.84FDD13 pKa = 3.65GQPTRR18 pKa = 11.84HH19 pKa = 6.29GIAPKK24 pKa = 10.24KK25 pKa = 9.93IFRR28 pKa = 11.84SKK30 pKa = 10.75YY31 pKa = 8.63IPTGLVPRR39 pKa = 11.84LKK41 pKa = 10.01YY42 pKa = 9.66WRR44 pKa = 11.84DD45 pKa = 3.29VPSRR49 pKa = 11.84AEE51 pKa = 3.55MSKK54 pKa = 10.65RR55 pKa = 11.84IGKK58 pKa = 9.31YY59 pKa = 9.85FEE61 pKa = 5.87DD62 pKa = 3.71EE63 pKa = 4.3FKK65 pKa = 10.67FYY67 pKa = 10.61PNEE70 pKa = 3.92EE71 pKa = 4.29KK72 pKa = 11.02LNEE75 pKa = 4.07AVNIVQEE82 pKa = 4.26KK83 pKa = 8.98WISHH87 pKa = 6.06YY88 pKa = 9.49GTSLNVTSVSEE99 pKa = 4.33SFRR102 pKa = 11.84TLPKK106 pKa = 9.45STSAGLPFKK115 pKa = 10.71SGCTKK120 pKa = 10.57YY121 pKa = 9.93EE122 pKa = 3.75ARR124 pKa = 11.84NKK126 pKa = 9.12MMRR129 pKa = 11.84FARR132 pKa = 11.84SQWDD136 pKa = 3.57RR137 pKa = 11.84VSKK140 pKa = 10.2EE141 pKa = 3.64LQLQVLPCRR150 pKa = 11.84LGARR154 pKa = 11.84CQLRR158 pKa = 11.84KK159 pKa = 9.98RR160 pKa = 11.84GEE162 pKa = 4.02NKK164 pKa = 9.47PRR166 pKa = 11.84LIWAYY171 pKa = 9.41PGYY174 pKa = 10.83LSIIEE179 pKa = 4.16NQYY182 pKa = 8.77LTAIKK187 pKa = 9.95KK188 pKa = 8.94VPPPNFIGWSTNWLDD203 pKa = 3.94GGKK206 pKa = 9.65SLNRR210 pKa = 11.84LLFGDD215 pKa = 3.26KK216 pKa = 8.38WTWQSIAQIDD226 pKa = 4.29FSSFDD231 pKa = 3.15ATVRR235 pKa = 11.84TEE237 pKa = 4.68LIFHH241 pKa = 7.16AFKK244 pKa = 10.69ILRR247 pKa = 11.84SLFDD251 pKa = 3.16LTRR254 pKa = 11.84TEE256 pKa = 5.79NIMLDD261 pKa = 3.47QLRR264 pKa = 11.84HH265 pKa = 4.97YY266 pKa = 9.77FINTPILFYY275 pKa = 11.29DD276 pKa = 4.11KK277 pKa = 10.58IIVKK281 pKa = 9.98NRR283 pKa = 11.84GIPSGSAFTQIIGTIVNMIACQYY306 pKa = 10.82ASLRR310 pKa = 11.84SRR312 pKa = 11.84DD313 pKa = 3.61YY314 pKa = 11.0NLRR317 pKa = 11.84IPFSCWLGDD326 pKa = 3.55DD327 pKa = 3.76SFLNFEE333 pKa = 4.29TALCRR338 pKa = 11.84QEE340 pKa = 5.24FEE342 pKa = 3.84YY343 pKa = 10.73DD344 pKa = 3.42YY345 pKa = 11.59LEE347 pKa = 4.26KK348 pKa = 10.76FKK350 pKa = 11.04EE351 pKa = 3.91LGLNVSIDD359 pKa = 3.34KK360 pKa = 9.0THH362 pKa = 5.42YY363 pKa = 4.91TTRR366 pKa = 11.84FIDD369 pKa = 3.83DD370 pKa = 3.52FEE372 pKa = 4.74VRR374 pKa = 11.84FKK376 pKa = 11.06GVRR379 pKa = 11.84PYY381 pKa = 11.53VKK383 pKa = 10.34FLGKK387 pKa = 10.44QIDD390 pKa = 3.6ILLDD394 pKa = 3.52LTFHH398 pKa = 7.21NDD400 pKa = 3.13LDD402 pKa = 4.16KK403 pKa = 11.6LDD405 pKa = 4.16AQMALPEE412 pKa = 4.58KK413 pKa = 9.71EE414 pKa = 4.61DD415 pKa = 3.43LSAYY419 pKa = 6.32EE420 pKa = 3.96TGVRR424 pKa = 11.84LIGLVWAYY432 pKa = 9.83GAHH435 pKa = 5.67YY436 pKa = 10.11DD437 pKa = 3.21IYY439 pKa = 11.33LRR441 pKa = 11.84ILKK444 pKa = 10.23VYY446 pKa = 10.63LSLKK450 pKa = 9.77LKK452 pKa = 10.71PEE454 pKa = 3.85FHH456 pKa = 6.04VQQLLSYY463 pKa = 9.24SEE465 pKa = 4.07KK466 pKa = 9.94PEE468 pKa = 3.68RR469 pKa = 11.84TKK471 pKa = 10.5RR472 pKa = 11.84YY473 pKa = 9.12QEE475 pKa = 4.03NFFSSMKK482 pKa = 10.19YY483 pKa = 9.78QLNLDD488 pKa = 3.74LDD490 pKa = 4.15IYY492 pKa = 11.44DD493 pKa = 4.95LLAFPKK499 pKa = 10.35FWDD502 pKa = 3.24VSNRR506 pKa = 11.84YY507 pKa = 8.86FGSKK511 pKa = 8.98YY512 pKa = 9.7EE513 pKa = 3.99RR514 pKa = 11.84LDD516 pKa = 3.6FRR518 pKa = 11.84SHH520 pKa = 7.25KK521 pKa = 10.48IYY523 pKa = 11.0GG524 pKa = 3.49
MM1 pKa = 7.41KK2 pKa = 10.09FVNIYY7 pKa = 9.6EE8 pKa = 4.09IQRR11 pKa = 11.84FDD13 pKa = 3.65GQPTRR18 pKa = 11.84HH19 pKa = 6.29GIAPKK24 pKa = 10.24KK25 pKa = 9.93IFRR28 pKa = 11.84SKK30 pKa = 10.75YY31 pKa = 8.63IPTGLVPRR39 pKa = 11.84LKK41 pKa = 10.01YY42 pKa = 9.66WRR44 pKa = 11.84DD45 pKa = 3.29VPSRR49 pKa = 11.84AEE51 pKa = 3.55MSKK54 pKa = 10.65RR55 pKa = 11.84IGKK58 pKa = 9.31YY59 pKa = 9.85FEE61 pKa = 5.87DD62 pKa = 3.71EE63 pKa = 4.3FKK65 pKa = 10.67FYY67 pKa = 10.61PNEE70 pKa = 3.92EE71 pKa = 4.29KK72 pKa = 11.02LNEE75 pKa = 4.07AVNIVQEE82 pKa = 4.26KK83 pKa = 8.98WISHH87 pKa = 6.06YY88 pKa = 9.49GTSLNVTSVSEE99 pKa = 4.33SFRR102 pKa = 11.84TLPKK106 pKa = 9.45STSAGLPFKK115 pKa = 10.71SGCTKK120 pKa = 10.57YY121 pKa = 9.93EE122 pKa = 3.75ARR124 pKa = 11.84NKK126 pKa = 9.12MMRR129 pKa = 11.84FARR132 pKa = 11.84SQWDD136 pKa = 3.57RR137 pKa = 11.84VSKK140 pKa = 10.2EE141 pKa = 3.64LQLQVLPCRR150 pKa = 11.84LGARR154 pKa = 11.84CQLRR158 pKa = 11.84KK159 pKa = 9.98RR160 pKa = 11.84GEE162 pKa = 4.02NKK164 pKa = 9.47PRR166 pKa = 11.84LIWAYY171 pKa = 9.41PGYY174 pKa = 10.83LSIIEE179 pKa = 4.16NQYY182 pKa = 8.77LTAIKK187 pKa = 9.95KK188 pKa = 8.94VPPPNFIGWSTNWLDD203 pKa = 3.94GGKK206 pKa = 9.65SLNRR210 pKa = 11.84LLFGDD215 pKa = 3.26KK216 pKa = 8.38WTWQSIAQIDD226 pKa = 4.29FSSFDD231 pKa = 3.15ATVRR235 pKa = 11.84TEE237 pKa = 4.68LIFHH241 pKa = 7.16AFKK244 pKa = 10.69ILRR247 pKa = 11.84SLFDD251 pKa = 3.16LTRR254 pKa = 11.84TEE256 pKa = 5.79NIMLDD261 pKa = 3.47QLRR264 pKa = 11.84HH265 pKa = 4.97YY266 pKa = 9.77FINTPILFYY275 pKa = 11.29DD276 pKa = 4.11KK277 pKa = 10.58IIVKK281 pKa = 9.98NRR283 pKa = 11.84GIPSGSAFTQIIGTIVNMIACQYY306 pKa = 10.82ASLRR310 pKa = 11.84SRR312 pKa = 11.84DD313 pKa = 3.61YY314 pKa = 11.0NLRR317 pKa = 11.84IPFSCWLGDD326 pKa = 3.55DD327 pKa = 3.76SFLNFEE333 pKa = 4.29TALCRR338 pKa = 11.84QEE340 pKa = 5.24FEE342 pKa = 3.84YY343 pKa = 10.73DD344 pKa = 3.42YY345 pKa = 11.59LEE347 pKa = 4.26KK348 pKa = 10.76FKK350 pKa = 11.04EE351 pKa = 3.91LGLNVSIDD359 pKa = 3.34KK360 pKa = 9.0THH362 pKa = 5.42YY363 pKa = 4.91TTRR366 pKa = 11.84FIDD369 pKa = 3.83DD370 pKa = 3.52FEE372 pKa = 4.74VRR374 pKa = 11.84FKK376 pKa = 11.06GVRR379 pKa = 11.84PYY381 pKa = 11.53VKK383 pKa = 10.34FLGKK387 pKa = 10.44QIDD390 pKa = 3.6ILLDD394 pKa = 3.52LTFHH398 pKa = 7.21NDD400 pKa = 3.13LDD402 pKa = 4.16KK403 pKa = 11.6LDD405 pKa = 4.16AQMALPEE412 pKa = 4.58KK413 pKa = 9.71EE414 pKa = 4.61DD415 pKa = 3.43LSAYY419 pKa = 6.32EE420 pKa = 3.96TGVRR424 pKa = 11.84LIGLVWAYY432 pKa = 9.83GAHH435 pKa = 5.67YY436 pKa = 10.11DD437 pKa = 3.21IYY439 pKa = 11.33LRR441 pKa = 11.84ILKK444 pKa = 10.23VYY446 pKa = 10.63LSLKK450 pKa = 9.77LKK452 pKa = 10.71PEE454 pKa = 3.85FHH456 pKa = 6.04VQQLLSYY463 pKa = 9.24SEE465 pKa = 4.07KK466 pKa = 9.94PEE468 pKa = 3.68RR469 pKa = 11.84TKK471 pKa = 10.5RR472 pKa = 11.84YY473 pKa = 9.12QEE475 pKa = 4.03NFFSSMKK482 pKa = 10.19YY483 pKa = 9.78QLNLDD488 pKa = 3.74LDD490 pKa = 4.15IYY492 pKa = 11.44DD493 pKa = 4.95LLAFPKK499 pKa = 10.35FWDD502 pKa = 3.24VSNRR506 pKa = 11.84YY507 pKa = 8.86FGSKK511 pKa = 8.98YY512 pKa = 9.7EE513 pKa = 3.99RR514 pKa = 11.84LDD516 pKa = 3.6FRR518 pKa = 11.84SHH520 pKa = 7.25KK521 pKa = 10.48IYY523 pKa = 11.0GG524 pKa = 3.49
Molecular weight: 61.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
843 |
319 |
524 |
421.5 |
49.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.915 ± 0.247 | 1.305 ± 0.139 |
5.694 ± 0.027 | 5.694 ± 0.139 |
5.457 ± 1.231 | 4.033 ± 0.809 |
2.491 ± 0.674 | 7.117 ± 0.049 |
7.117 ± 0.616 | 10.558 ± 0.113 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.898 ± 0.323 | 4.508 ± 0.269 |
4.745 ± 0.476 | 3.559 ± 0.225 |
6.762 ± 0.261 | 7.236 ± 0.319 |
5.694 ± 0.97 | 5.219 ± 0.889 |
1.779 ± 0.279 | 5.219 ± 0.441 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
