
Auxenochlorella protothecoides (Green microalga) (Chlorella protothecoides)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Trebouxiophyceae; Chlorellales; Chlorellaceae; Auxenochlorella
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
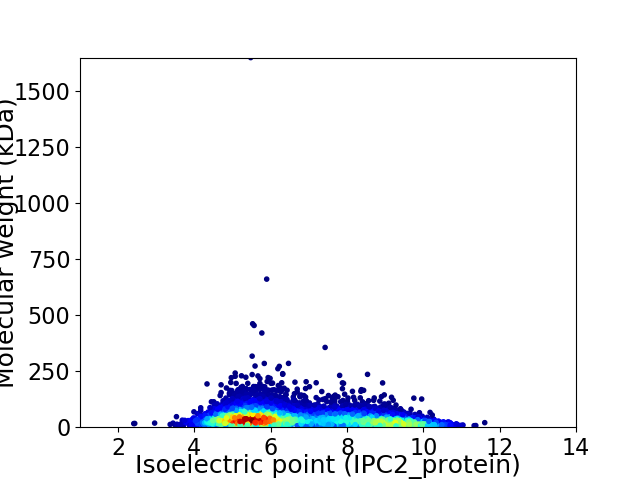
Virtual 2D-PAGE plot for 7001 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A087ST42|A0A087ST42_AUXPR VAMP-like protein YKT61 OS=Auxenochlorella protothecoides OX=3075 GN=APUTEX25_001748 PE=3 SV=1
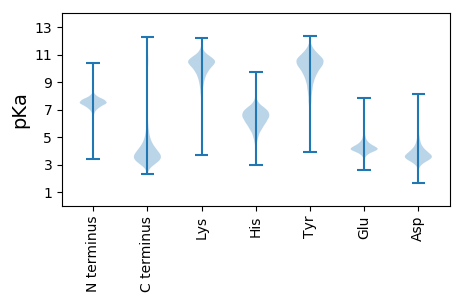
MM1 pKa = 7.68KK2 pKa = 10.05FAQLVIGPAGCGKK15 pKa = 8.66STYY18 pKa = 10.36CEE20 pKa = 4.02TIKK23 pKa = 10.75AHH25 pKa = 7.23CDD27 pKa = 3.12TIGRR31 pKa = 11.84SVHH34 pKa = 5.76VVNLDD39 pKa = 3.35PAAEE43 pKa = 4.08HH44 pKa = 6.29FNYY47 pKa = 9.7PVSIDD52 pKa = 3.51VRR54 pKa = 11.84DD55 pKa = 4.95LITLDD60 pKa = 4.19DD61 pKa = 3.93VAEE64 pKa = 4.24EE65 pKa = 4.7LDD67 pKa = 3.7LGPNGGLLYY76 pKa = 10.95CMEE79 pKa = 4.44FLEE82 pKa = 5.96DD83 pKa = 5.32NLSDD87 pKa = 3.37WLEE90 pKa = 4.23EE91 pKa = 3.85EE92 pKa = 4.14LQGYY96 pKa = 9.44GEE98 pKa = 4.83DD99 pKa = 4.65DD100 pKa = 4.02YY101 pKa = 12.11LLFDD105 pKa = 4.49CPGQIEE111 pKa = 5.09LYY113 pKa = 9.42SHH115 pKa = 6.48VSVFRR120 pKa = 11.84TFTEE124 pKa = 4.16FLKK127 pKa = 10.66RR128 pKa = 11.84DD129 pKa = 3.18GWNICAVYY137 pKa = 10.75CMDD140 pKa = 3.59SQFVAEE146 pKa = 4.58PPKK149 pKa = 10.57FIAGCLSALSAMVQLEE165 pKa = 4.31LPHH168 pKa = 8.23VNMLTKK174 pKa = 10.35VDD176 pKa = 4.27LVQDD180 pKa = 3.68KK181 pKa = 11.15DD182 pKa = 3.82SLEE185 pKa = 4.48SYY187 pKa = 10.01LFPDD191 pKa = 3.75ALEE194 pKa = 4.38LRR196 pKa = 11.84HH197 pKa = 6.58ALGEE201 pKa = 4.37STGPRR206 pKa = 11.84FSALNDD212 pKa = 3.6VIVNLLDD219 pKa = 3.45EE220 pKa = 4.56FAMVSFTPLDD230 pKa = 3.57IGDD233 pKa = 3.98EE234 pKa = 4.19EE235 pKa = 5.1SIEE238 pKa = 4.17TALFQVDD245 pKa = 3.54SAIQYY250 pKa = 10.98GEE252 pKa = 4.19DD253 pKa = 3.02QDD255 pKa = 4.66VRR257 pKa = 11.84QKK259 pKa = 11.21EE260 pKa = 4.28YY261 pKa = 11.32GDD263 pKa = 4.15FPDD266 pKa = 4.44EE267 pKa = 3.73EE268 pKa = 5.29APAHH272 pKa = 5.39EE273 pKa = 5.66AIDD276 pKa = 3.81FF277 pKa = 4.19
MM1 pKa = 7.68KK2 pKa = 10.05FAQLVIGPAGCGKK15 pKa = 8.66STYY18 pKa = 10.36CEE20 pKa = 4.02TIKK23 pKa = 10.75AHH25 pKa = 7.23CDD27 pKa = 3.12TIGRR31 pKa = 11.84SVHH34 pKa = 5.76VVNLDD39 pKa = 3.35PAAEE43 pKa = 4.08HH44 pKa = 6.29FNYY47 pKa = 9.7PVSIDD52 pKa = 3.51VRR54 pKa = 11.84DD55 pKa = 4.95LITLDD60 pKa = 4.19DD61 pKa = 3.93VAEE64 pKa = 4.24EE65 pKa = 4.7LDD67 pKa = 3.7LGPNGGLLYY76 pKa = 10.95CMEE79 pKa = 4.44FLEE82 pKa = 5.96DD83 pKa = 5.32NLSDD87 pKa = 3.37WLEE90 pKa = 4.23EE91 pKa = 3.85EE92 pKa = 4.14LQGYY96 pKa = 9.44GEE98 pKa = 4.83DD99 pKa = 4.65DD100 pKa = 4.02YY101 pKa = 12.11LLFDD105 pKa = 4.49CPGQIEE111 pKa = 5.09LYY113 pKa = 9.42SHH115 pKa = 6.48VSVFRR120 pKa = 11.84TFTEE124 pKa = 4.16FLKK127 pKa = 10.66RR128 pKa = 11.84DD129 pKa = 3.18GWNICAVYY137 pKa = 10.75CMDD140 pKa = 3.59SQFVAEE146 pKa = 4.58PPKK149 pKa = 10.57FIAGCLSALSAMVQLEE165 pKa = 4.31LPHH168 pKa = 8.23VNMLTKK174 pKa = 10.35VDD176 pKa = 4.27LVQDD180 pKa = 3.68KK181 pKa = 11.15DD182 pKa = 3.82SLEE185 pKa = 4.48SYY187 pKa = 10.01LFPDD191 pKa = 3.75ALEE194 pKa = 4.38LRR196 pKa = 11.84HH197 pKa = 6.58ALGEE201 pKa = 4.37STGPRR206 pKa = 11.84FSALNDD212 pKa = 3.6VIVNLLDD219 pKa = 3.45EE220 pKa = 4.56FAMVSFTPLDD230 pKa = 3.57IGDD233 pKa = 3.98EE234 pKa = 4.19EE235 pKa = 5.1SIEE238 pKa = 4.17TALFQVDD245 pKa = 3.54SAIQYY250 pKa = 10.98GEE252 pKa = 4.19DD253 pKa = 3.02QDD255 pKa = 4.66VRR257 pKa = 11.84QKK259 pKa = 11.21EE260 pKa = 4.28YY261 pKa = 11.32GDD263 pKa = 4.15FPDD266 pKa = 4.44EE267 pKa = 3.73EE268 pKa = 5.29APAHH272 pKa = 5.39EE273 pKa = 5.66AIDD276 pKa = 3.81FF277 pKa = 4.19
Molecular weight: 30.97 kDa
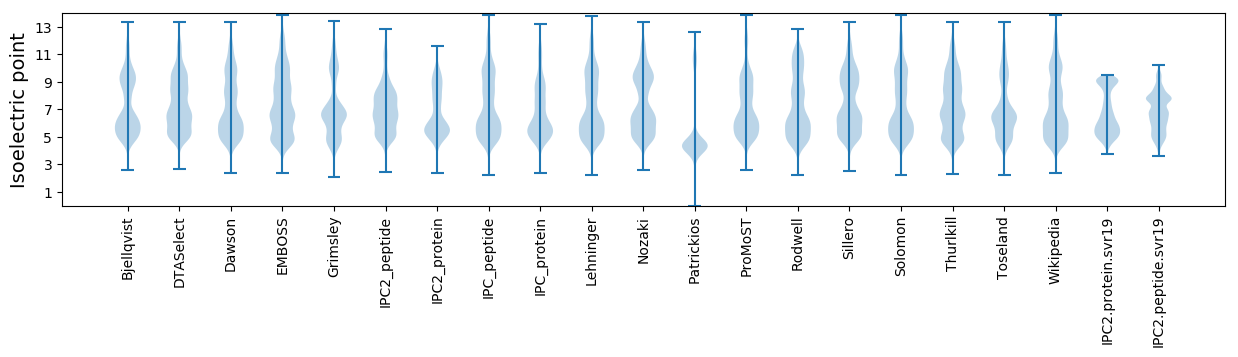
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A087SIX0|A0A087SIX0_AUXPR DUF1995 domain-containing protein OS=Auxenochlorella protothecoides OX=3075 GN=APUTEX25_004025 PE=4 SV=1
MM1 pKa = 7.37ARR3 pKa = 11.84LAATHH8 pKa = 6.6LAQRR12 pKa = 11.84GRR14 pKa = 11.84RR15 pKa = 11.84SSLRR19 pKa = 11.84VWISPTLTSGSRR31 pKa = 11.84HH32 pKa = 5.36AFWRR36 pKa = 11.84TSWRR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84VLGRR47 pKa = 11.84GRR49 pKa = 11.84RR50 pKa = 11.84PPLPPWSRR58 pKa = 11.84AAA60 pKa = 3.72
MM1 pKa = 7.37ARR3 pKa = 11.84LAATHH8 pKa = 6.6LAQRR12 pKa = 11.84GRR14 pKa = 11.84RR15 pKa = 11.84SSLRR19 pKa = 11.84VWISPTLTSGSRR31 pKa = 11.84HH32 pKa = 5.36AFWRR36 pKa = 11.84TSWRR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84VLGRR47 pKa = 11.84GRR49 pKa = 11.84RR50 pKa = 11.84PPLPPWSRR58 pKa = 11.84AAA60 pKa = 3.72
Molecular weight: 6.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2763953 |
49 |
16440 |
394.8 |
42.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.692 ± 0.061 | 1.513 ± 0.012 |
4.951 ± 0.018 | 5.862 ± 0.03 |
2.922 ± 0.017 | 8.823 ± 0.03 |
2.311 ± 0.014 | 2.96 ± 0.021 |
3.066 ± 0.028 | 10.716 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.953 ± 0.013 | 2.021 ± 0.016 |
6.257 ± 0.026 | 3.662 ± 0.018 |
7.136 ± 0.028 | 6.492 ± 0.023 |
4.881 ± 0.019 | 6.974 ± 0.024 |
1.48 ± 0.012 | 2.059 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
