
Citricoccus sp. SGAir0253
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Citricoccus; unclassified Citricoccus
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
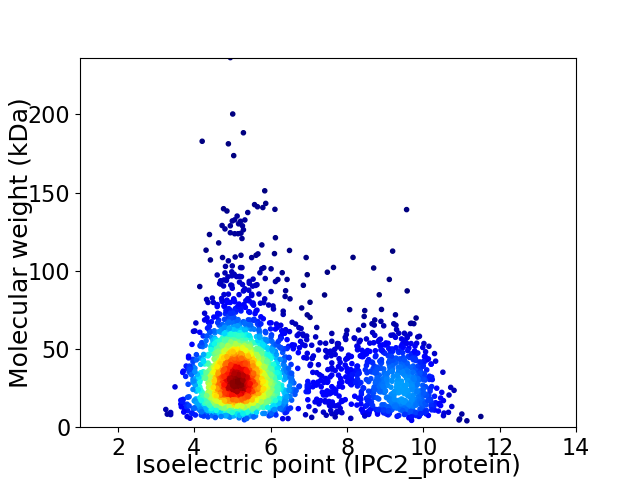
Virtual 2D-PAGE plot for 2944 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P9JAH5|A0A4P9JAH5_9MICC Glycosyltransferase family 9 protein OS=Citricoccus sp. SGAir0253 OX=2567881 GN=E7744_07695 PE=4 SV=1
MM1 pKa = 6.76KK2 pKa = 9.18TARR5 pKa = 11.84LEE7 pKa = 4.36SPAVRR12 pKa = 11.84AAALAAVTVPFLAACGGGADD32 pKa = 4.11GGEE35 pKa = 4.27TLTFAAIPAEE45 pKa = 4.26SSASLEE51 pKa = 3.91SDD53 pKa = 3.52YY54 pKa = 12.01SNITALIEE62 pKa = 4.01QEE64 pKa = 4.08TGVDD68 pKa = 3.61VEE70 pKa = 4.49FQNASDD76 pKa = 3.83YY77 pKa = 11.44AAVIEE82 pKa = 4.37GQRR85 pKa = 11.84AGQIDD90 pKa = 3.76IASYY94 pKa = 11.09GPFSYY99 pKa = 10.89VIAKK103 pKa = 10.39DD104 pKa = 3.17SGVNIEE110 pKa = 4.6PVAAPTSDD118 pKa = 3.36QDD120 pKa = 3.49KK121 pKa = 10.48QPAYY125 pKa = 9.64TSLAYY130 pKa = 10.42VRR132 pKa = 11.84ADD134 pKa = 4.2SDD136 pKa = 3.69IQDD139 pKa = 3.99LADD142 pKa = 3.95LEE144 pKa = 4.55GKK146 pKa = 9.57KK147 pKa = 10.26VCFVDD152 pKa = 3.61AASTSGYY159 pKa = 9.41LVPMKK164 pKa = 10.72GLLDD168 pKa = 3.76EE169 pKa = 6.2GIDD172 pKa = 4.07LNTEE176 pKa = 3.99TEE178 pKa = 3.91AVLAGGHH185 pKa = 6.27DD186 pKa = 4.03ASLLSLDD193 pKa = 5.23AGSCDD198 pKa = 3.49AAFAHH203 pKa = 6.43DD204 pKa = 3.9TMLNTLVTSGQVDD217 pKa = 3.9EE218 pKa = 5.44GALKK222 pKa = 9.93PVWEE226 pKa = 4.48SAPITEE232 pKa = 5.64DD233 pKa = 4.27PIALNMDD240 pKa = 4.26TIEE243 pKa = 4.94PEE245 pKa = 3.86LAEE248 pKa = 4.27QISTVIRR255 pKa = 11.84EE256 pKa = 4.31KK257 pKa = 10.98ANKK260 pKa = 8.25PALVEE265 pKa = 4.11AGICASEE272 pKa = 4.22EE273 pKa = 4.12DD274 pKa = 4.04CVLPEE279 pKa = 4.38EE280 pKa = 4.56IEE282 pKa = 4.16YY283 pKa = 10.43GYY285 pKa = 11.23LPVDD289 pKa = 4.5DD290 pKa = 5.65SDD292 pKa = 3.77FDD294 pKa = 4.84AIRR297 pKa = 11.84EE298 pKa = 4.05ICAATDD304 pKa = 3.09ADD306 pKa = 3.9ACHH309 pKa = 6.61SVGG312 pKa = 4.61
MM1 pKa = 6.76KK2 pKa = 9.18TARR5 pKa = 11.84LEE7 pKa = 4.36SPAVRR12 pKa = 11.84AAALAAVTVPFLAACGGGADD32 pKa = 4.11GGEE35 pKa = 4.27TLTFAAIPAEE45 pKa = 4.26SSASLEE51 pKa = 3.91SDD53 pKa = 3.52YY54 pKa = 12.01SNITALIEE62 pKa = 4.01QEE64 pKa = 4.08TGVDD68 pKa = 3.61VEE70 pKa = 4.49FQNASDD76 pKa = 3.83YY77 pKa = 11.44AAVIEE82 pKa = 4.37GQRR85 pKa = 11.84AGQIDD90 pKa = 3.76IASYY94 pKa = 11.09GPFSYY99 pKa = 10.89VIAKK103 pKa = 10.39DD104 pKa = 3.17SGVNIEE110 pKa = 4.6PVAAPTSDD118 pKa = 3.36QDD120 pKa = 3.49KK121 pKa = 10.48QPAYY125 pKa = 9.64TSLAYY130 pKa = 10.42VRR132 pKa = 11.84ADD134 pKa = 4.2SDD136 pKa = 3.69IQDD139 pKa = 3.99LADD142 pKa = 3.95LEE144 pKa = 4.55GKK146 pKa = 9.57KK147 pKa = 10.26VCFVDD152 pKa = 3.61AASTSGYY159 pKa = 9.41LVPMKK164 pKa = 10.72GLLDD168 pKa = 3.76EE169 pKa = 6.2GIDD172 pKa = 4.07LNTEE176 pKa = 3.99TEE178 pKa = 3.91AVLAGGHH185 pKa = 6.27DD186 pKa = 4.03ASLLSLDD193 pKa = 5.23AGSCDD198 pKa = 3.49AAFAHH203 pKa = 6.43DD204 pKa = 3.9TMLNTLVTSGQVDD217 pKa = 3.9EE218 pKa = 5.44GALKK222 pKa = 9.93PVWEE226 pKa = 4.48SAPITEE232 pKa = 5.64DD233 pKa = 4.27PIALNMDD240 pKa = 4.26TIEE243 pKa = 4.94PEE245 pKa = 3.86LAEE248 pKa = 4.27QISTVIRR255 pKa = 11.84EE256 pKa = 4.31KK257 pKa = 10.98ANKK260 pKa = 8.25PALVEE265 pKa = 4.11AGICASEE272 pKa = 4.22EE273 pKa = 4.12DD274 pKa = 4.04CVLPEE279 pKa = 4.38EE280 pKa = 4.56IEE282 pKa = 4.16YY283 pKa = 10.43GYY285 pKa = 11.23LPVDD289 pKa = 4.5DD290 pKa = 5.65SDD292 pKa = 3.77FDD294 pKa = 4.84AIRR297 pKa = 11.84EE298 pKa = 4.05ICAATDD304 pKa = 3.09ADD306 pKa = 3.9ACHH309 pKa = 6.61SVGG312 pKa = 4.61
Molecular weight: 32.49 kDa
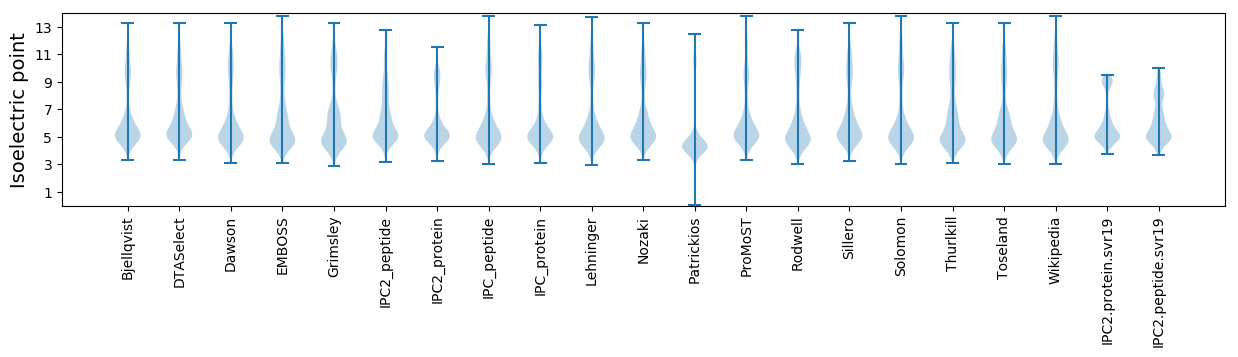
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P9JBT8|A0A4P9JBT8_9MICC ABC transporter substrate-binding protein OS=Citricoccus sp. SGAir0253 OX=2567881 GN=E7744_09900 PE=4 SV=1
MM1 pKa = 7.41RR2 pKa = 11.84WARR5 pKa = 11.84PSSSAPRR12 pKa = 11.84TPSPAPAGPASRR24 pKa = 11.84SPTPARR30 pKa = 11.84TARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84PRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84PVRR47 pKa = 11.84PRR49 pKa = 11.84PAPTVRR55 pKa = 11.84RR56 pKa = 11.84GAARR60 pKa = 11.84GGG62 pKa = 3.48
MM1 pKa = 7.41RR2 pKa = 11.84WARR5 pKa = 11.84PSSSAPRR12 pKa = 11.84TPSPAPAGPASRR24 pKa = 11.84SPTPARR30 pKa = 11.84TARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84PRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84PVRR47 pKa = 11.84PRR49 pKa = 11.84PAPTVRR55 pKa = 11.84RR56 pKa = 11.84GAARR60 pKa = 11.84GGG62 pKa = 3.48
Molecular weight: 6.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1013340 |
32 |
2154 |
344.2 |
36.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.018 ± 0.072 | 0.594 ± 0.011 |
5.842 ± 0.042 | 5.979 ± 0.042 |
2.739 ± 0.029 | 9.903 ± 0.047 |
2.256 ± 0.021 | 3.27 ± 0.031 |
1.605 ± 0.032 | 10.113 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.881 ± 0.02 | 1.701 ± 0.022 |
6.178 ± 0.04 | 2.793 ± 0.028 |
7.923 ± 0.054 | 4.92 ± 0.031 |
5.971 ± 0.033 | 8.895 ± 0.046 |
1.509 ± 0.02 | 1.913 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
