
Microbacterium phage Phinky
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Squashvirus; unclassified Squashvirus
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
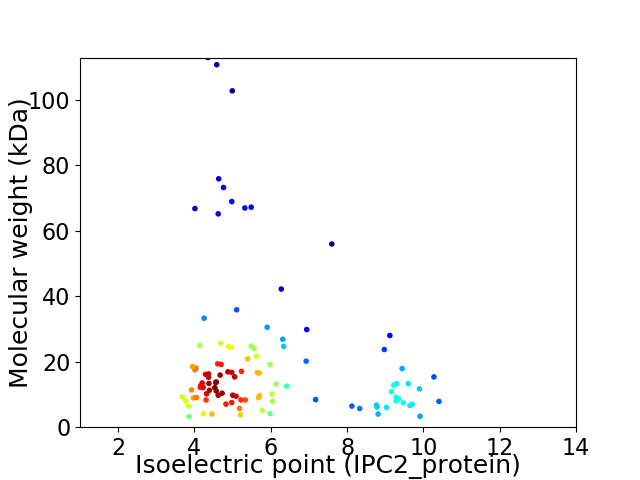
Virtual 2D-PAGE plot for 106 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
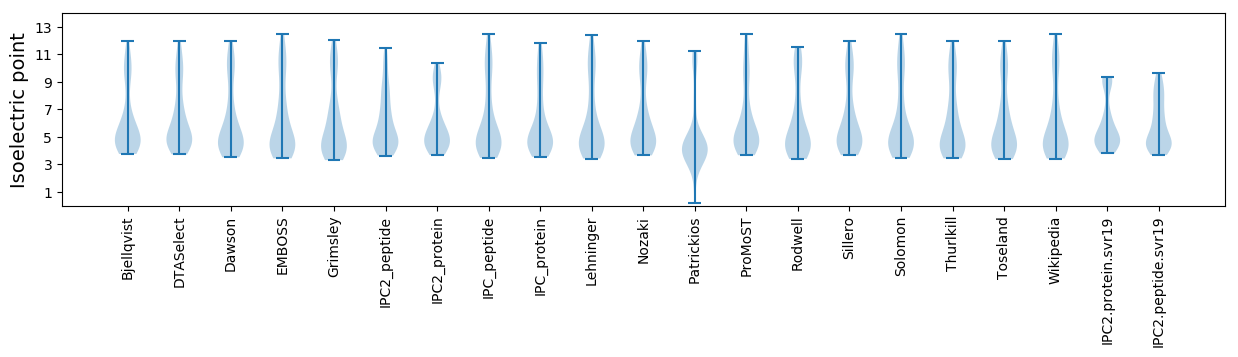
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7G8LEA1|A0A7G8LEA1_9CAUD Uncharacterized protein OS=Microbacterium phage Phinky OX=2762418 GN=82 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.55KK3 pKa = 10.56FIACAATAAIATLGLTALAVPASATTLDD31 pKa = 3.55SFGICVPTEE40 pKa = 3.97DD41 pKa = 3.36TTQTIVHH48 pKa = 6.75PAVGTPTIEE57 pKa = 4.26VNNPDD62 pKa = 3.93YY63 pKa = 11.36VPAVPGTPGVPAIGEE78 pKa = 4.08PTIEE82 pKa = 4.47IPNPAYY88 pKa = 10.81VPGVPAVPGTPAVGEE103 pKa = 4.12EE104 pKa = 4.51TILVPNPGYY113 pKa = 10.78VPAVPAVPGTPAVGEE128 pKa = 4.07PTIEE132 pKa = 4.25VPNPDD137 pKa = 3.92YY138 pKa = 10.96VAPTYY143 pKa = 10.65TPGFWQDD150 pKa = 3.02VPAVGTPTIIVTKK163 pKa = 9.43PNPDD167 pKa = 3.41YY168 pKa = 11.12VPAVAEE174 pKa = 4.06IPEE177 pKa = 4.26VSHH180 pKa = 6.44TDD182 pKa = 3.65YY183 pKa = 10.77VAKK186 pKa = 10.29RR187 pKa = 11.84YY188 pKa = 10.6VIDD191 pKa = 3.51QAAQPEE197 pKa = 4.59VTKK200 pKa = 8.5TQWRR204 pKa = 11.84YY205 pKa = 9.14VKK207 pKa = 10.52NGGYY211 pKa = 10.6GEE213 pKa = 4.38VWLDD217 pKa = 3.35SNAEE221 pKa = 4.09HH222 pKa = 6.74KK223 pKa = 11.1VKK225 pKa = 10.73INGYY229 pKa = 8.05WYY231 pKa = 10.19EE232 pKa = 4.13RR233 pKa = 11.84TNKK236 pKa = 8.78TKK238 pKa = 10.19TVVVTPAIPEE248 pKa = 4.13RR249 pKa = 11.84GHH251 pKa = 6.56WEE253 pKa = 3.64QQSFHH258 pKa = 7.31EE259 pKa = 4.39YY260 pKa = 9.92PGAGWTILSEE270 pKa = 4.56SKK272 pKa = 10.61HH273 pKa = 4.16VTQEE277 pKa = 4.03YY278 pKa = 10.23VPGSPAQGSEE288 pKa = 4.17TIDD291 pKa = 3.42VEE293 pKa = 4.45EE294 pKa = 4.84ANPDD298 pKa = 3.61YY299 pKa = 11.47VPATTVWVPPVYY311 pKa = 10.31TPAQGTKK318 pKa = 9.78TIAKK322 pKa = 9.13DD323 pKa = 3.44NPDD326 pKa = 3.65YY327 pKa = 11.51VPATEE332 pKa = 5.27EE333 pKa = 4.05IPGTPAVGEE342 pKa = 4.36PEE344 pKa = 4.75LEE346 pKa = 4.46VPNPAYY352 pKa = 9.83IPATEE357 pKa = 4.25GTPEE361 pKa = 3.67IPAVGEE367 pKa = 3.85EE368 pKa = 4.86TITIDD373 pKa = 3.22NPAYY377 pKa = 9.65IPAVPAVDD385 pKa = 5.15EE386 pKa = 4.47IPAVGEE392 pKa = 3.67PTIEE396 pKa = 4.14VEE398 pKa = 4.2NPDD401 pKa = 4.4YY402 pKa = 10.92IPEE405 pKa = 3.99RR406 pKa = 11.84TEE408 pKa = 3.99IITIPGTEE416 pKa = 4.58CPPVIPEE423 pKa = 4.07EE424 pKa = 4.31PEE426 pKa = 3.9EE427 pKa = 4.12PDD429 pKa = 3.32YY430 pKa = 11.28TLLDD434 pKa = 3.6GSISVDD440 pKa = 3.47CVAGTVTFRR449 pKa = 11.84GSNGTDD455 pKa = 3.04EE456 pKa = 4.51TVEE459 pKa = 4.0FATGFDD465 pKa = 3.7SDD467 pKa = 3.72GDD469 pKa = 4.13GAVDD473 pKa = 4.58FGDD476 pKa = 4.6GFIAPADD483 pKa = 3.61GAGQFVYY490 pKa = 10.1TFAEE494 pKa = 4.15LAGNEE499 pKa = 4.4SLTAHH504 pKa = 6.6LFIWGEE510 pKa = 4.03GWDD513 pKa = 3.88VAHH516 pKa = 6.47TVATSDD522 pKa = 3.19EE523 pKa = 4.32FSPVVCEE530 pKa = 4.2TPTEE534 pKa = 4.28PEE536 pKa = 4.01EE537 pKa = 4.1PTTPEE542 pKa = 3.86EE543 pKa = 4.1PTTPEE548 pKa = 3.86EE549 pKa = 4.1PTTPEE554 pKa = 3.98EE555 pKa = 4.03PTVPEE560 pKa = 4.53EE561 pKa = 4.08PTVPTTPVTPAEE573 pKa = 4.04PVAPVVNPAPAAPAAGAARR592 pKa = 11.84AAKK595 pKa = 10.38APVAAEE601 pKa = 4.32ADD603 pKa = 3.81TLAQTGADD611 pKa = 3.49VAFGAVGAAGVALLAGAGLLLHH633 pKa = 6.94RR634 pKa = 11.84RR635 pKa = 11.84RR636 pKa = 11.84HH637 pKa = 4.64TSS639 pKa = 2.67
MM1 pKa = 7.54KK2 pKa = 10.55KK3 pKa = 10.56FIACAATAAIATLGLTALAVPASATTLDD31 pKa = 3.55SFGICVPTEE40 pKa = 3.97DD41 pKa = 3.36TTQTIVHH48 pKa = 6.75PAVGTPTIEE57 pKa = 4.26VNNPDD62 pKa = 3.93YY63 pKa = 11.36VPAVPGTPGVPAIGEE78 pKa = 4.08PTIEE82 pKa = 4.47IPNPAYY88 pKa = 10.81VPGVPAVPGTPAVGEE103 pKa = 4.12EE104 pKa = 4.51TILVPNPGYY113 pKa = 10.78VPAVPAVPGTPAVGEE128 pKa = 4.07PTIEE132 pKa = 4.25VPNPDD137 pKa = 3.92YY138 pKa = 10.96VAPTYY143 pKa = 10.65TPGFWQDD150 pKa = 3.02VPAVGTPTIIVTKK163 pKa = 9.43PNPDD167 pKa = 3.41YY168 pKa = 11.12VPAVAEE174 pKa = 4.06IPEE177 pKa = 4.26VSHH180 pKa = 6.44TDD182 pKa = 3.65YY183 pKa = 10.77VAKK186 pKa = 10.29RR187 pKa = 11.84YY188 pKa = 10.6VIDD191 pKa = 3.51QAAQPEE197 pKa = 4.59VTKK200 pKa = 8.5TQWRR204 pKa = 11.84YY205 pKa = 9.14VKK207 pKa = 10.52NGGYY211 pKa = 10.6GEE213 pKa = 4.38VWLDD217 pKa = 3.35SNAEE221 pKa = 4.09HH222 pKa = 6.74KK223 pKa = 11.1VKK225 pKa = 10.73INGYY229 pKa = 8.05WYY231 pKa = 10.19EE232 pKa = 4.13RR233 pKa = 11.84TNKK236 pKa = 8.78TKK238 pKa = 10.19TVVVTPAIPEE248 pKa = 4.13RR249 pKa = 11.84GHH251 pKa = 6.56WEE253 pKa = 3.64QQSFHH258 pKa = 7.31EE259 pKa = 4.39YY260 pKa = 9.92PGAGWTILSEE270 pKa = 4.56SKK272 pKa = 10.61HH273 pKa = 4.16VTQEE277 pKa = 4.03YY278 pKa = 10.23VPGSPAQGSEE288 pKa = 4.17TIDD291 pKa = 3.42VEE293 pKa = 4.45EE294 pKa = 4.84ANPDD298 pKa = 3.61YY299 pKa = 11.47VPATTVWVPPVYY311 pKa = 10.31TPAQGTKK318 pKa = 9.78TIAKK322 pKa = 9.13DD323 pKa = 3.44NPDD326 pKa = 3.65YY327 pKa = 11.51VPATEE332 pKa = 5.27EE333 pKa = 4.05IPGTPAVGEE342 pKa = 4.36PEE344 pKa = 4.75LEE346 pKa = 4.46VPNPAYY352 pKa = 9.83IPATEE357 pKa = 4.25GTPEE361 pKa = 3.67IPAVGEE367 pKa = 3.85EE368 pKa = 4.86TITIDD373 pKa = 3.22NPAYY377 pKa = 9.65IPAVPAVDD385 pKa = 5.15EE386 pKa = 4.47IPAVGEE392 pKa = 3.67PTIEE396 pKa = 4.14VEE398 pKa = 4.2NPDD401 pKa = 4.4YY402 pKa = 10.92IPEE405 pKa = 3.99RR406 pKa = 11.84TEE408 pKa = 3.99IITIPGTEE416 pKa = 4.58CPPVIPEE423 pKa = 4.07EE424 pKa = 4.31PEE426 pKa = 3.9EE427 pKa = 4.12PDD429 pKa = 3.32YY430 pKa = 11.28TLLDD434 pKa = 3.6GSISVDD440 pKa = 3.47CVAGTVTFRR449 pKa = 11.84GSNGTDD455 pKa = 3.04EE456 pKa = 4.51TVEE459 pKa = 4.0FATGFDD465 pKa = 3.7SDD467 pKa = 3.72GDD469 pKa = 4.13GAVDD473 pKa = 4.58FGDD476 pKa = 4.6GFIAPADD483 pKa = 3.61GAGQFVYY490 pKa = 10.1TFAEE494 pKa = 4.15LAGNEE499 pKa = 4.4SLTAHH504 pKa = 6.6LFIWGEE510 pKa = 4.03GWDD513 pKa = 3.88VAHH516 pKa = 6.47TVATSDD522 pKa = 3.19EE523 pKa = 4.32FSPVVCEE530 pKa = 4.2TPTEE534 pKa = 4.28PEE536 pKa = 4.01EE537 pKa = 4.1PTTPEE542 pKa = 3.86EE543 pKa = 4.1PTTPEE548 pKa = 3.86EE549 pKa = 4.1PTTPEE554 pKa = 3.98EE555 pKa = 4.03PTVPEE560 pKa = 4.53EE561 pKa = 4.08PTVPTTPVTPAEE573 pKa = 4.04PVAPVVNPAPAAPAAGAARR592 pKa = 11.84AAKK595 pKa = 10.38APVAAEE601 pKa = 4.32ADD603 pKa = 3.81TLAQTGADD611 pKa = 3.49VAFGAVGAAGVALLAGAGLLLHH633 pKa = 6.94RR634 pKa = 11.84RR635 pKa = 11.84RR636 pKa = 11.84HH637 pKa = 4.64TSS639 pKa = 2.67
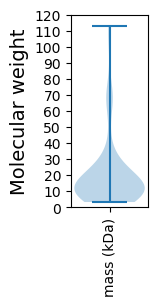
Molecular weight: 66.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7G8LE38|A0A7G8LE38_9CAUD Helix-turn-helix DNA binding domain protein OS=Microbacterium phage Phinky OX=2762418 GN=18 PE=4 SV=1
MM1 pKa = 7.88AKK3 pKa = 9.46RR4 pKa = 11.84DD5 pKa = 3.69RR6 pKa = 11.84PRR8 pKa = 11.84YY9 pKa = 8.45IPPRR13 pKa = 11.84LGQRR17 pKa = 11.84DD18 pKa = 3.89FARR21 pKa = 11.84TAMVPRR27 pKa = 11.84WFTRR31 pKa = 11.84EE32 pKa = 3.31AVAAFRR38 pKa = 11.84LTPYY42 pKa = 10.58DD43 pKa = 3.69FAAFTVIADD52 pKa = 3.75NLDD55 pKa = 3.25SHH57 pKa = 6.83GISTTAMTLIATRR70 pKa = 11.84GGMSEE75 pKa = 4.11GGAAKK80 pKa = 10.13AVNRR84 pKa = 11.84LIAVDD89 pKa = 5.7LIHH92 pKa = 6.88EE93 pKa = 4.98LDD95 pKa = 3.74PRR97 pKa = 11.84RR98 pKa = 11.84KK99 pKa = 8.95GHH101 pKa = 5.26IMRR104 pKa = 11.84YY105 pKa = 9.79GIAPDD110 pKa = 3.73IPWTEE115 pKa = 4.06TQRR118 pKa = 3.61
MM1 pKa = 7.88AKK3 pKa = 9.46RR4 pKa = 11.84DD5 pKa = 3.69RR6 pKa = 11.84PRR8 pKa = 11.84YY9 pKa = 8.45IPPRR13 pKa = 11.84LGQRR17 pKa = 11.84DD18 pKa = 3.89FARR21 pKa = 11.84TAMVPRR27 pKa = 11.84WFTRR31 pKa = 11.84EE32 pKa = 3.31AVAAFRR38 pKa = 11.84LTPYY42 pKa = 10.58DD43 pKa = 3.69FAAFTVIADD52 pKa = 3.75NLDD55 pKa = 3.25SHH57 pKa = 6.83GISTTAMTLIATRR70 pKa = 11.84GGMSEE75 pKa = 4.11GGAAKK80 pKa = 10.13AVNRR84 pKa = 11.84LIAVDD89 pKa = 5.7LIHH92 pKa = 6.88EE93 pKa = 4.98LDD95 pKa = 3.74PRR97 pKa = 11.84RR98 pKa = 11.84KK99 pKa = 8.95GHH101 pKa = 5.26IMRR104 pKa = 11.84YY105 pKa = 9.79GIAPDD110 pKa = 3.73IPWTEE115 pKa = 4.06TQRR118 pKa = 3.61
Molecular weight: 13.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
20058 |
30 |
1055 |
189.2 |
20.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.447 ± 0.363 | 0.853 ± 0.126 |
6.342 ± 0.204 | 7.005 ± 0.344 |
2.797 ± 0.131 | 8.575 ± 0.235 |
1.855 ± 0.172 | 4.906 ± 0.142 |
2.587 ± 0.203 | 7.394 ± 0.348 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.243 ± 0.138 | 2.991 ± 0.188 |
5.564 ± 0.384 | 3.724 ± 0.284 |
6.97 ± 0.347 | 5.788 ± 0.343 |
7.214 ± 0.474 | 7.344 ± 0.208 |
1.944 ± 0.157 | 2.458 ± 0.16 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
