
Gulosibacter sp. 10
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Gulosibacter; unclassified Gulosibacter
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
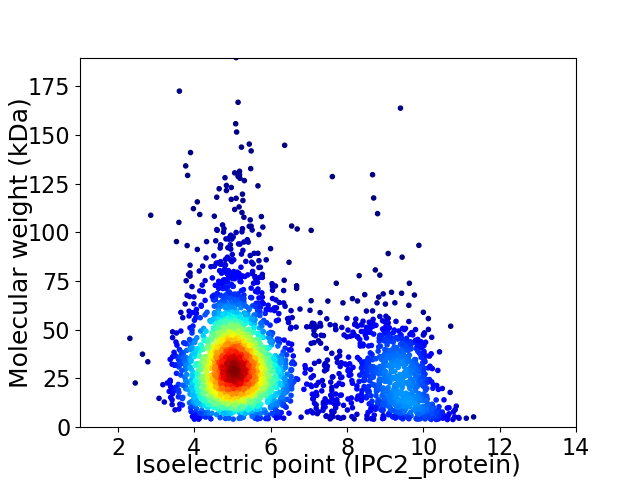
Virtual 2D-PAGE plot for 3335 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1R4FLH9|A0A1R4FLH9_9MICO Hydantoin racemase OS=Gulosibacter sp. 10 OX=1255570 GN=FM112_04910 PE=4 SV=1
MM1 pKa = 7.6NKK3 pKa = 10.12RR4 pKa = 11.84IVSGVALVAAFGLVLSGCARR24 pKa = 11.84ADD26 pKa = 3.5EE27 pKa = 4.67AGGGGVAEE35 pKa = 5.24DD36 pKa = 3.99GGQVEE41 pKa = 4.94HH42 pKa = 7.08LNAAEE47 pKa = 3.94NQMNYY52 pKa = 9.69VDD54 pKa = 4.8EE55 pKa = 6.1DD56 pKa = 4.25GLGHH60 pKa = 7.19GGTLRR65 pKa = 11.84LYY67 pKa = 10.96NNAYY71 pKa = 7.98PANFNFNTVEE81 pKa = 4.23GYY83 pKa = 9.29EE84 pKa = 4.19KK85 pKa = 9.81NTNEE89 pKa = 3.82IMEE92 pKa = 4.49AVWPQLYY99 pKa = 10.3AYY101 pKa = 7.77TADD104 pKa = 3.65GQVIPNDD111 pKa = 4.02LYY113 pKa = 8.86TQRR116 pKa = 11.84IEE118 pKa = 4.3QVSEE122 pKa = 3.84EE123 pKa = 4.07PFAYY127 pKa = 9.91EE128 pKa = 3.67IEE130 pKa = 4.35LVEE133 pKa = 4.26GLTWSDD139 pKa = 3.42GTPLDD144 pKa = 3.49WTSVKK149 pKa = 11.05NNMEE153 pKa = 4.4AYY155 pKa = 10.05QNEE158 pKa = 4.25EE159 pKa = 4.12FNVVSRR165 pKa = 11.84DD166 pKa = 3.32GYY168 pKa = 11.33DD169 pKa = 3.28RR170 pKa = 11.84IEE172 pKa = 4.41EE173 pKa = 4.19VTQGDD178 pKa = 3.94NEE180 pKa = 4.13QTAVITFKK188 pKa = 10.8EE189 pKa = 4.54GEE191 pKa = 4.55VYY193 pKa = 10.9ADD195 pKa = 3.96WIGLSGVMPDD205 pKa = 4.04ALVEE209 pKa = 4.21SAEE212 pKa = 4.14EE213 pKa = 4.63FNTGWVDD220 pKa = 3.83GPKK223 pKa = 8.91VTAGPYY229 pKa = 9.14TIEE232 pKa = 4.04NADD235 pKa = 3.98RR236 pKa = 11.84ANQVVTLVPDD246 pKa = 4.11EE247 pKa = 4.39NWTGARR253 pKa = 11.84DD254 pKa = 3.62AKK256 pKa = 10.63LDD258 pKa = 3.65RR259 pKa = 11.84LSFQTIEE266 pKa = 4.62DD267 pKa = 3.82PSAAATAFNNGQLDD281 pKa = 4.21VIDD284 pKa = 4.95ASAEE288 pKa = 3.98AIYY291 pKa = 10.71SVVIDD296 pKa = 3.89TVDD299 pKa = 3.22NGDD302 pKa = 3.47EE303 pKa = 4.26FEE305 pKa = 4.48IRR307 pKa = 11.84TAAGPDD313 pKa = 3.13WSHH316 pKa = 5.95FTLNGGEE323 pKa = 4.37GNVLSDD329 pKa = 3.77PNLRR333 pKa = 11.84EE334 pKa = 3.85AFFYY338 pKa = 10.71AINRR342 pKa = 11.84ADD344 pKa = 3.81IFMALNGTMPYY355 pKa = 10.28PEE357 pKa = 5.57GIEE360 pKa = 3.84NDD362 pKa = 3.41LLGNHH367 pKa = 6.16MLMSNQDD374 pKa = 3.31GYY376 pKa = 11.22EE377 pKa = 4.07SHH379 pKa = 7.03AGEE382 pKa = 4.99YY383 pKa = 8.76GTGDD387 pKa = 3.42ADD389 pKa = 4.04RR390 pKa = 11.84AKK392 pKa = 10.78EE393 pKa = 4.0LIEE396 pKa = 3.9EE397 pKa = 4.61AGWEE401 pKa = 4.0LGDD404 pKa = 4.68DD405 pKa = 4.02GFYY408 pKa = 10.91QKK410 pKa = 10.77DD411 pKa = 3.92GEE413 pKa = 4.42VLEE416 pKa = 3.96IRR418 pKa = 11.84YY419 pKa = 9.57VYY421 pKa = 10.7NAGSAVNGTIAPIATEE437 pKa = 4.01NLQAAGIKK445 pKa = 9.22LTVEE449 pKa = 3.96QVPPTDD455 pKa = 4.31LFSQYY460 pKa = 10.64VIPGDD465 pKa = 3.51YY466 pKa = 10.39DD467 pKa = 2.9ITMFGWSGNPFVTSSVNSWLSDD489 pKa = 3.39GEE491 pKa = 4.47QNFTNIGSEE500 pKa = 4.17EE501 pKa = 4.57LDD503 pKa = 3.77DD504 pKa = 6.17LLNQLTNEE512 pKa = 4.14ADD514 pKa = 3.23VDD516 pKa = 4.01KK517 pKa = 10.71QTDD520 pKa = 3.92LLNQIDD526 pKa = 3.82EE527 pKa = 4.77ALWGLHH533 pKa = 6.44SNLPLYY539 pKa = 10.34QSYY542 pKa = 11.26DD543 pKa = 4.21FIATPVDD550 pKa = 3.53LANYY554 pKa = 7.41GAPGFANIIDD564 pKa = 3.86WSMVGYY570 pKa = 10.85VDD572 pKa = 5.33GSPHH576 pKa = 7.74LEE578 pKa = 4.09GG579 pKa = 5.32
MM1 pKa = 7.6NKK3 pKa = 10.12RR4 pKa = 11.84IVSGVALVAAFGLVLSGCARR24 pKa = 11.84ADD26 pKa = 3.5EE27 pKa = 4.67AGGGGVAEE35 pKa = 5.24DD36 pKa = 3.99GGQVEE41 pKa = 4.94HH42 pKa = 7.08LNAAEE47 pKa = 3.94NQMNYY52 pKa = 9.69VDD54 pKa = 4.8EE55 pKa = 6.1DD56 pKa = 4.25GLGHH60 pKa = 7.19GGTLRR65 pKa = 11.84LYY67 pKa = 10.96NNAYY71 pKa = 7.98PANFNFNTVEE81 pKa = 4.23GYY83 pKa = 9.29EE84 pKa = 4.19KK85 pKa = 9.81NTNEE89 pKa = 3.82IMEE92 pKa = 4.49AVWPQLYY99 pKa = 10.3AYY101 pKa = 7.77TADD104 pKa = 3.65GQVIPNDD111 pKa = 4.02LYY113 pKa = 8.86TQRR116 pKa = 11.84IEE118 pKa = 4.3QVSEE122 pKa = 3.84EE123 pKa = 4.07PFAYY127 pKa = 9.91EE128 pKa = 3.67IEE130 pKa = 4.35LVEE133 pKa = 4.26GLTWSDD139 pKa = 3.42GTPLDD144 pKa = 3.49WTSVKK149 pKa = 11.05NNMEE153 pKa = 4.4AYY155 pKa = 10.05QNEE158 pKa = 4.25EE159 pKa = 4.12FNVVSRR165 pKa = 11.84DD166 pKa = 3.32GYY168 pKa = 11.33DD169 pKa = 3.28RR170 pKa = 11.84IEE172 pKa = 4.41EE173 pKa = 4.19VTQGDD178 pKa = 3.94NEE180 pKa = 4.13QTAVITFKK188 pKa = 10.8EE189 pKa = 4.54GEE191 pKa = 4.55VYY193 pKa = 10.9ADD195 pKa = 3.96WIGLSGVMPDD205 pKa = 4.04ALVEE209 pKa = 4.21SAEE212 pKa = 4.14EE213 pKa = 4.63FNTGWVDD220 pKa = 3.83GPKK223 pKa = 8.91VTAGPYY229 pKa = 9.14TIEE232 pKa = 4.04NADD235 pKa = 3.98RR236 pKa = 11.84ANQVVTLVPDD246 pKa = 4.11EE247 pKa = 4.39NWTGARR253 pKa = 11.84DD254 pKa = 3.62AKK256 pKa = 10.63LDD258 pKa = 3.65RR259 pKa = 11.84LSFQTIEE266 pKa = 4.62DD267 pKa = 3.82PSAAATAFNNGQLDD281 pKa = 4.21VIDD284 pKa = 4.95ASAEE288 pKa = 3.98AIYY291 pKa = 10.71SVVIDD296 pKa = 3.89TVDD299 pKa = 3.22NGDD302 pKa = 3.47EE303 pKa = 4.26FEE305 pKa = 4.48IRR307 pKa = 11.84TAAGPDD313 pKa = 3.13WSHH316 pKa = 5.95FTLNGGEE323 pKa = 4.37GNVLSDD329 pKa = 3.77PNLRR333 pKa = 11.84EE334 pKa = 3.85AFFYY338 pKa = 10.71AINRR342 pKa = 11.84ADD344 pKa = 3.81IFMALNGTMPYY355 pKa = 10.28PEE357 pKa = 5.57GIEE360 pKa = 3.84NDD362 pKa = 3.41LLGNHH367 pKa = 6.16MLMSNQDD374 pKa = 3.31GYY376 pKa = 11.22EE377 pKa = 4.07SHH379 pKa = 7.03AGEE382 pKa = 4.99YY383 pKa = 8.76GTGDD387 pKa = 3.42ADD389 pKa = 4.04RR390 pKa = 11.84AKK392 pKa = 10.78EE393 pKa = 4.0LIEE396 pKa = 3.9EE397 pKa = 4.61AGWEE401 pKa = 4.0LGDD404 pKa = 4.68DD405 pKa = 4.02GFYY408 pKa = 10.91QKK410 pKa = 10.77DD411 pKa = 3.92GEE413 pKa = 4.42VLEE416 pKa = 3.96IRR418 pKa = 11.84YY419 pKa = 9.57VYY421 pKa = 10.7NAGSAVNGTIAPIATEE437 pKa = 4.01NLQAAGIKK445 pKa = 9.22LTVEE449 pKa = 3.96QVPPTDD455 pKa = 4.31LFSQYY460 pKa = 10.64VIPGDD465 pKa = 3.51YY466 pKa = 10.39DD467 pKa = 2.9ITMFGWSGNPFVTSSVNSWLSDD489 pKa = 3.39GEE491 pKa = 4.47QNFTNIGSEE500 pKa = 4.17EE501 pKa = 4.57LDD503 pKa = 3.77DD504 pKa = 6.17LLNQLTNEE512 pKa = 4.14ADD514 pKa = 3.23VDD516 pKa = 4.01KK517 pKa = 10.71QTDD520 pKa = 3.92LLNQIDD526 pKa = 3.82EE527 pKa = 4.77ALWGLHH533 pKa = 6.44SNLPLYY539 pKa = 10.34QSYY542 pKa = 11.26DD543 pKa = 4.21FIATPVDD550 pKa = 3.53LANYY554 pKa = 7.41GAPGFANIIDD564 pKa = 3.86WSMVGYY570 pKa = 10.85VDD572 pKa = 5.33GSPHH576 pKa = 7.74LEE578 pKa = 4.09GG579 pKa = 5.32
Molecular weight: 63.28 kDa
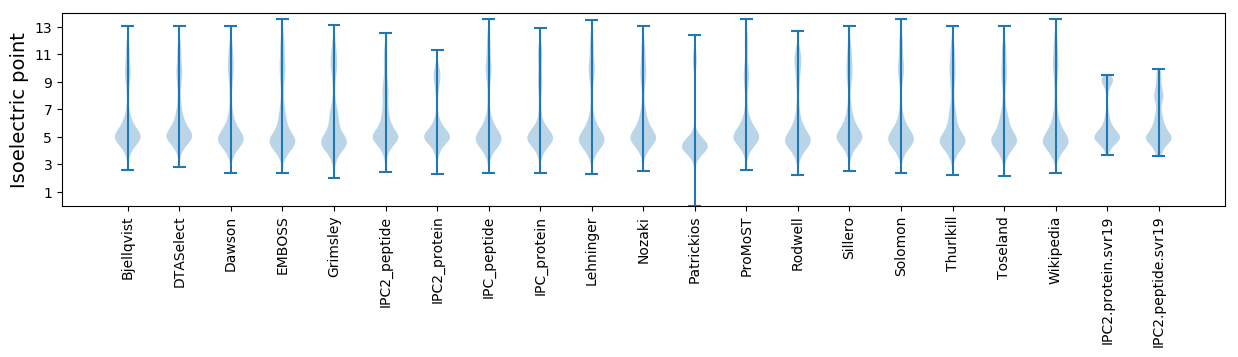
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1R4G5I3|A0A1R4G5I3_9MICO RNA-binding protein Jag OS=Gulosibacter sp. 10 OX=1255570 GN=FM112_09140 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84KK16 pKa = 8.24HH17 pKa = 4.59GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSSRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 9.71GRR40 pKa = 11.84KK41 pKa = 8.78VLSAA45 pKa = 4.05
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84KK16 pKa = 8.24HH17 pKa = 4.59GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSSRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 9.71GRR40 pKa = 11.84KK41 pKa = 8.78VLSAA45 pKa = 4.05
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1064179 |
37 |
1803 |
319.1 |
34.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.199 ± 0.06 | 0.566 ± 0.01 |
5.818 ± 0.037 | 7.134 ± 0.046 |
3.192 ± 0.025 | 9.259 ± 0.044 |
1.993 ± 0.02 | 4.604 ± 0.034 |
1.819 ± 0.028 | 10.228 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.91 ± 0.018 | 1.915 ± 0.023 |
5.41 ± 0.033 | 2.823 ± 0.03 |
7.948 ± 0.057 | 5.401 ± 0.028 |
5.283 ± 0.03 | 8.125 ± 0.034 |
1.395 ± 0.016 | 1.98 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
