
Tobacco virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
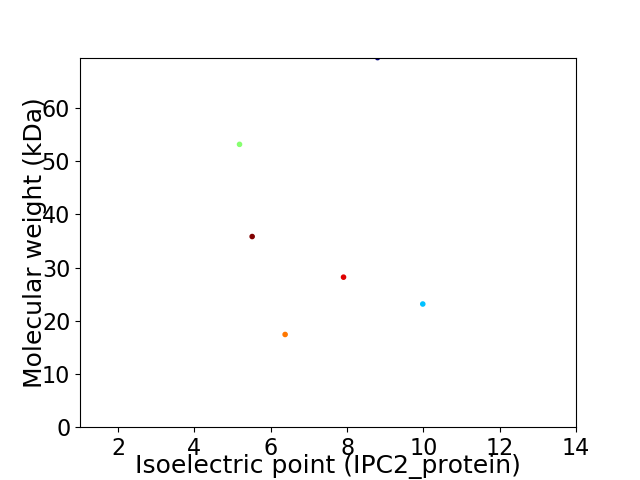
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
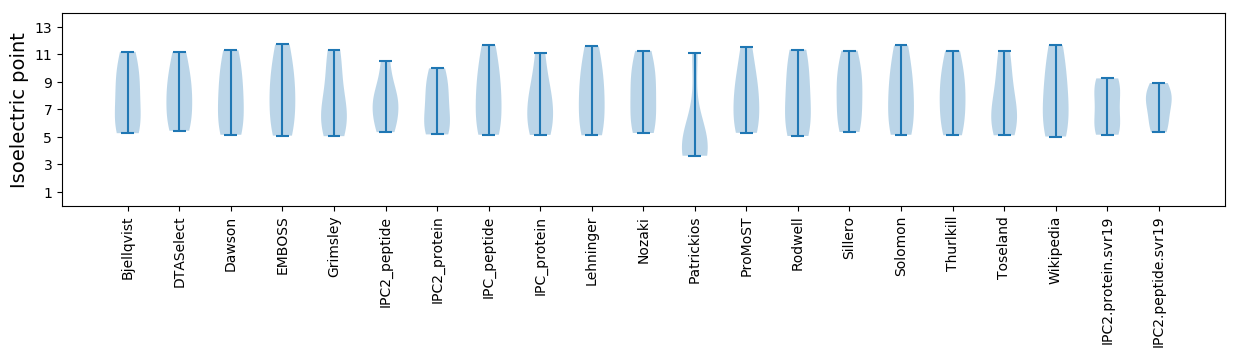
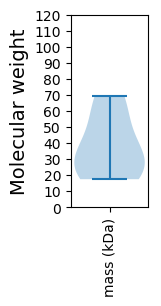
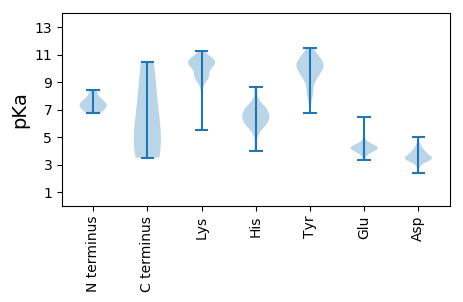
Protein with the lowest isoelectric point:
>tr|A0A1V0JHW2|A0A1V0JHW2_9LUTE RNA-directed RNA polymerase OS=Tobacco virus 2 OX=1972995 PE=4 SV=1
MM1 pKa = 7.23LTIQARR7 pKa = 11.84EE8 pKa = 3.85NDD10 pKa = 3.67DD11 pKa = 3.94QIILGSLGSQRR22 pKa = 11.84MKK24 pKa = 10.93YY25 pKa = 9.9IEE27 pKa = 5.51DD28 pKa = 3.36EE29 pKa = 4.18NQNYY33 pKa = 8.21TKK35 pKa = 10.51FSSEE39 pKa = 4.59YY40 pKa = 8.59YY41 pKa = 9.92SQSSMQAVPMYY52 pKa = 9.89YY53 pKa = 10.3FNVPKK58 pKa = 10.36GQWSVDD64 pKa = 3.39ISCEE68 pKa = 4.18GYY70 pKa = 10.21QPTSSTSDD78 pKa = 3.55PNRR81 pKa = 11.84GRR83 pKa = 11.84SDD85 pKa = 3.4GMIAYY90 pKa = 10.35SNADD94 pKa = 2.79SDD96 pKa = 4.02YY97 pKa = 9.5WNVGEE102 pKa = 4.53ADD104 pKa = 3.49GVKK107 pKa = 9.88ISKK110 pKa = 9.59LRR112 pKa = 11.84NDD114 pKa = 2.86NTYY117 pKa = 10.53RR118 pKa = 11.84QGHH121 pKa = 6.79PEE123 pKa = 4.05LEE125 pKa = 4.1INSCHH130 pKa = 5.84FRR132 pKa = 11.84EE133 pKa = 4.68GQLLEE138 pKa = 4.26RR139 pKa = 11.84DD140 pKa = 3.63ATISFHH146 pKa = 8.31VEE148 pKa = 3.32APTDD152 pKa = 3.4GRR154 pKa = 11.84FFLVGPAIQKK164 pKa = 5.49TAKK167 pKa = 9.33YY168 pKa = 9.73NYY170 pKa = 8.05TISYY174 pKa = 10.17GDD176 pKa = 3.06WTDD179 pKa = 3.7RR180 pKa = 11.84DD181 pKa = 3.91MEE183 pKa = 4.99LGLITVVLDD192 pKa = 3.29EE193 pKa = 4.77HH194 pKa = 8.61LEE196 pKa = 4.34GTGSANRR203 pKa = 11.84VRR205 pKa = 11.84RR206 pKa = 11.84PPRR209 pKa = 11.84EE210 pKa = 3.32GHH212 pKa = 6.13TYY214 pKa = 8.54MASPHH219 pKa = 5.77EE220 pKa = 4.38PEE222 pKa = 4.9GKK224 pKa = 9.47PVGNKK229 pKa = 9.3PRR231 pKa = 11.84DD232 pKa = 3.45EE233 pKa = 4.47TPIQTQEE240 pKa = 4.03RR241 pKa = 11.84QPDD244 pKa = 3.65QTPSDD249 pKa = 4.17DD250 pKa = 3.7VSDD253 pKa = 4.08AGSVNSGGPTEE264 pKa = 4.13SLRR267 pKa = 11.84LEE269 pKa = 4.24FGVNSDD275 pKa = 3.13STYY278 pKa = 11.27DD279 pKa = 3.5ATVDD283 pKa = 3.44GTDD286 pKa = 3.38WPRR289 pKa = 11.84IPPPRR294 pKa = 11.84HH295 pKa = 5.24PPEE298 pKa = 4.29PRR300 pKa = 11.84VSGNSRR306 pKa = 11.84TVTDD310 pKa = 5.0FSSKK314 pKa = 11.18ADD316 pKa = 4.0LLEE319 pKa = 4.27NWDD322 pKa = 4.58AEE324 pKa = 4.39HH325 pKa = 7.23FDD327 pKa = 3.6PGYY330 pKa = 10.6SKK332 pKa = 11.2EE333 pKa = 4.24DD334 pKa = 3.24VAAATIIAHH343 pKa = 6.99GSIQDD348 pKa = 3.53GRR350 pKa = 11.84SMLEE354 pKa = 3.66KK355 pKa = 10.35RR356 pKa = 11.84EE357 pKa = 4.03EE358 pKa = 3.98NVKK361 pKa = 10.85NKK363 pKa = 8.99TSSWKK368 pKa = 9.93PPSLKK373 pKa = 10.48AVSPAIAKK381 pKa = 9.51LRR383 pKa = 11.84SIRR386 pKa = 11.84KK387 pKa = 7.69SQPLEE392 pKa = 4.04GGTLNKK398 pKa = 10.38DD399 pKa = 3.0ATDD402 pKa = 4.12GVSSIGSGSLTGGTLKK418 pKa = 10.59RR419 pKa = 11.84KK420 pKa = 8.53ATIEE424 pKa = 3.84EE425 pKa = 4.38RR426 pKa = 11.84LLQTLTTEE434 pKa = 3.67QRR436 pKa = 11.84LWYY439 pKa = 10.26EE440 pKa = 4.08NFKK443 pKa = 9.32KK444 pKa = 9.31TNPPAATQWLFEE456 pKa = 4.28YY457 pKa = 10.49QPPPQVDD464 pKa = 3.24RR465 pKa = 11.84NIAEE469 pKa = 4.33KK470 pKa = 10.39PFQGRR475 pKa = 11.84KK476 pKa = 8.39
MM1 pKa = 7.23LTIQARR7 pKa = 11.84EE8 pKa = 3.85NDD10 pKa = 3.67DD11 pKa = 3.94QIILGSLGSQRR22 pKa = 11.84MKK24 pKa = 10.93YY25 pKa = 9.9IEE27 pKa = 5.51DD28 pKa = 3.36EE29 pKa = 4.18NQNYY33 pKa = 8.21TKK35 pKa = 10.51FSSEE39 pKa = 4.59YY40 pKa = 8.59YY41 pKa = 9.92SQSSMQAVPMYY52 pKa = 9.89YY53 pKa = 10.3FNVPKK58 pKa = 10.36GQWSVDD64 pKa = 3.39ISCEE68 pKa = 4.18GYY70 pKa = 10.21QPTSSTSDD78 pKa = 3.55PNRR81 pKa = 11.84GRR83 pKa = 11.84SDD85 pKa = 3.4GMIAYY90 pKa = 10.35SNADD94 pKa = 2.79SDD96 pKa = 4.02YY97 pKa = 9.5WNVGEE102 pKa = 4.53ADD104 pKa = 3.49GVKK107 pKa = 9.88ISKK110 pKa = 9.59LRR112 pKa = 11.84NDD114 pKa = 2.86NTYY117 pKa = 10.53RR118 pKa = 11.84QGHH121 pKa = 6.79PEE123 pKa = 4.05LEE125 pKa = 4.1INSCHH130 pKa = 5.84FRR132 pKa = 11.84EE133 pKa = 4.68GQLLEE138 pKa = 4.26RR139 pKa = 11.84DD140 pKa = 3.63ATISFHH146 pKa = 8.31VEE148 pKa = 3.32APTDD152 pKa = 3.4GRR154 pKa = 11.84FFLVGPAIQKK164 pKa = 5.49TAKK167 pKa = 9.33YY168 pKa = 9.73NYY170 pKa = 8.05TISYY174 pKa = 10.17GDD176 pKa = 3.06WTDD179 pKa = 3.7RR180 pKa = 11.84DD181 pKa = 3.91MEE183 pKa = 4.99LGLITVVLDD192 pKa = 3.29EE193 pKa = 4.77HH194 pKa = 8.61LEE196 pKa = 4.34GTGSANRR203 pKa = 11.84VRR205 pKa = 11.84RR206 pKa = 11.84PPRR209 pKa = 11.84EE210 pKa = 3.32GHH212 pKa = 6.13TYY214 pKa = 8.54MASPHH219 pKa = 5.77EE220 pKa = 4.38PEE222 pKa = 4.9GKK224 pKa = 9.47PVGNKK229 pKa = 9.3PRR231 pKa = 11.84DD232 pKa = 3.45EE233 pKa = 4.47TPIQTQEE240 pKa = 4.03RR241 pKa = 11.84QPDD244 pKa = 3.65QTPSDD249 pKa = 4.17DD250 pKa = 3.7VSDD253 pKa = 4.08AGSVNSGGPTEE264 pKa = 4.13SLRR267 pKa = 11.84LEE269 pKa = 4.24FGVNSDD275 pKa = 3.13STYY278 pKa = 11.27DD279 pKa = 3.5ATVDD283 pKa = 3.44GTDD286 pKa = 3.38WPRR289 pKa = 11.84IPPPRR294 pKa = 11.84HH295 pKa = 5.24PPEE298 pKa = 4.29PRR300 pKa = 11.84VSGNSRR306 pKa = 11.84TVTDD310 pKa = 5.0FSSKK314 pKa = 11.18ADD316 pKa = 4.0LLEE319 pKa = 4.27NWDD322 pKa = 4.58AEE324 pKa = 4.39HH325 pKa = 7.23FDD327 pKa = 3.6PGYY330 pKa = 10.6SKK332 pKa = 11.2EE333 pKa = 4.24DD334 pKa = 3.24VAAATIIAHH343 pKa = 6.99GSIQDD348 pKa = 3.53GRR350 pKa = 11.84SMLEE354 pKa = 3.66KK355 pKa = 10.35RR356 pKa = 11.84EE357 pKa = 4.03EE358 pKa = 3.98NVKK361 pKa = 10.85NKK363 pKa = 8.99TSSWKK368 pKa = 9.93PPSLKK373 pKa = 10.48AVSPAIAKK381 pKa = 9.51LRR383 pKa = 11.84SIRR386 pKa = 11.84KK387 pKa = 7.69SQPLEE392 pKa = 4.04GGTLNKK398 pKa = 10.38DD399 pKa = 3.0ATDD402 pKa = 4.12GVSSIGSGSLTGGTLKK418 pKa = 10.59RR419 pKa = 11.84KK420 pKa = 8.53ATIEE424 pKa = 3.84EE425 pKa = 4.38RR426 pKa = 11.84LLQTLTTEE434 pKa = 3.67QRR436 pKa = 11.84LWYY439 pKa = 10.26EE440 pKa = 4.08NFKK443 pKa = 9.32KK444 pKa = 9.31TNPPAATQWLFEE456 pKa = 4.28YY457 pKa = 10.49QPPPQVDD464 pKa = 3.24RR465 pKa = 11.84NIAEE469 pKa = 4.33KK470 pKa = 10.39PFQGRR475 pKa = 11.84KK476 pKa = 8.39
Molecular weight: 53.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V0JHV8|A0A1V0JHV8_9LUTE Movement protein OS=Tobacco virus 2 OX=1972995 PE=3 SV=1
MM1 pKa = 6.72STVVVKK7 pKa = 10.83GNVNGGVQQPRR18 pKa = 11.84MRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84QSLRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84ANRR31 pKa = 11.84VQPVVMVTAPGQPRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84GGNRR55 pKa = 11.84RR56 pKa = 11.84SRR58 pKa = 11.84RR59 pKa = 11.84TGVPRR64 pKa = 11.84GRR66 pKa = 11.84GSSEE70 pKa = 3.73TFVFTKK76 pKa = 10.79DD77 pKa = 3.28NLVGNTQGSFTFGPSLSDD95 pKa = 3.42CPAFKK100 pKa = 10.76DD101 pKa = 4.47GILKK105 pKa = 10.29AYY107 pKa = 9.81HH108 pKa = 6.59EE109 pKa = 4.33YY110 pKa = 10.69KK111 pKa = 9.21ITSILLQFVSEE122 pKa = 4.33ASSTSSGSIAYY133 pKa = 9.43EE134 pKa = 3.86LDD136 pKa = 3.29PHH138 pKa = 6.86CKK140 pKa = 9.78VSSLQSYY147 pKa = 7.92VNKK150 pKa = 10.02FQITKK155 pKa = 10.26GGAKK159 pKa = 9.43TYY161 pKa = 7.27QARR164 pKa = 11.84MINGVEE170 pKa = 3.88WHH172 pKa = 7.28DD173 pKa = 4.24SSEE176 pKa = 4.13DD177 pKa = 3.25QCRR180 pKa = 11.84ILWKK184 pKa = 10.94GNGKK188 pKa = 10.24SSDD191 pKa = 3.53SAGSFRR197 pKa = 11.84VTIKK201 pKa = 10.78VALQNPKK208 pKa = 10.46
MM1 pKa = 6.72STVVVKK7 pKa = 10.83GNVNGGVQQPRR18 pKa = 11.84MRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84QSLRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84ANRR31 pKa = 11.84VQPVVMVTAPGQPRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84GGNRR55 pKa = 11.84RR56 pKa = 11.84SRR58 pKa = 11.84RR59 pKa = 11.84TGVPRR64 pKa = 11.84GRR66 pKa = 11.84GSSEE70 pKa = 3.73TFVFTKK76 pKa = 10.79DD77 pKa = 3.28NLVGNTQGSFTFGPSLSDD95 pKa = 3.42CPAFKK100 pKa = 10.76DD101 pKa = 4.47GILKK105 pKa = 10.29AYY107 pKa = 9.81HH108 pKa = 6.59EE109 pKa = 4.33YY110 pKa = 10.69KK111 pKa = 9.21ITSILLQFVSEE122 pKa = 4.33ASSTSSGSIAYY133 pKa = 9.43EE134 pKa = 3.86LDD136 pKa = 3.29PHH138 pKa = 6.86CKK140 pKa = 9.78VSSLQSYY147 pKa = 7.92VNKK150 pKa = 10.02FQITKK155 pKa = 10.26GGAKK159 pKa = 9.43TYY161 pKa = 7.27QARR164 pKa = 11.84MINGVEE170 pKa = 3.88WHH172 pKa = 7.28DD173 pKa = 4.24SSEE176 pKa = 4.13DD177 pKa = 3.25QCRR180 pKa = 11.84ILWKK184 pKa = 10.94GNGKK188 pKa = 10.24SSDD191 pKa = 3.53SAGSFRR197 pKa = 11.84VTIKK201 pKa = 10.78VALQNPKK208 pKa = 10.46
Molecular weight: 23.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2039 |
156 |
636 |
339.8 |
37.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.357 ± 0.903 | 1.667 ± 0.379 |
3.973 ± 0.863 | 6.376 ± 0.801 |
3.58 ± 0.4 | 6.719 ± 0.559 |
1.913 ± 0.283 | 4.708 ± 0.462 |
6.13 ± 0.811 | 8.092 ± 1.301 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.962 ± 0.27 | 4.414 ± 0.277 |
6.229 ± 0.718 | 4.169 ± 0.419 |
5.885 ± 0.982 | 9.564 ± 0.661 |
6.278 ± 0.37 | 5.64 ± 0.406 |
1.717 ± 0.119 | 3.629 ± 0.289 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
