
Aphanomyces stellatus
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Saprolegniales; Saprolegniaceae; Aphanomyces
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
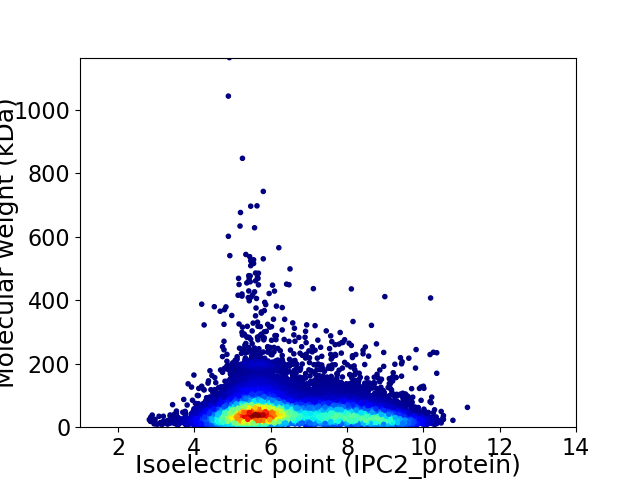
Virtual 2D-PAGE plot for 20738 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A485KTB5|A0A485KTB5_9STRA Aste57867_11406 protein OS=Aphanomyces stellatus OX=120398 GN=Aste57867_11406 PE=4 SV=1
MM1 pKa = 7.3NALHH5 pKa = 6.81HH6 pKa = 7.03ADD8 pKa = 3.82CAANGAPALDD18 pKa = 3.99EE19 pKa = 5.06DD20 pKa = 4.35EE21 pKa = 5.27QILATFSRR29 pKa = 11.84TQMYY33 pKa = 8.61TNTYY37 pKa = 10.1GPDD40 pKa = 3.33EE41 pKa = 5.29DD42 pKa = 6.07DD43 pKa = 3.75NGTMEE48 pKa = 4.27HH49 pKa = 6.59KK50 pKa = 10.73GVGEE54 pKa = 4.31LYY56 pKa = 9.55VTTTRR61 pKa = 11.84IAWVFGTNWEE71 pKa = 4.13HH72 pKa = 7.55DD73 pKa = 3.42AGAVVGYY80 pKa = 9.71AWDD83 pKa = 3.5MTYY86 pKa = 11.29LSLHH90 pKa = 6.68AISRR94 pKa = 11.84DD95 pKa = 2.9ISSFRR100 pKa = 11.84FPCLYY105 pKa = 10.39CQLDD109 pKa = 3.64VEE111 pKa = 5.1DD112 pKa = 4.64EE113 pKa = 4.29VNEE116 pKa = 3.89IRR118 pKa = 11.84FVPQEE123 pKa = 4.07PEE125 pKa = 3.96KK126 pKa = 10.48EE127 pKa = 4.04LQAMFDD133 pKa = 3.77AFSASAALNPDD144 pKa = 3.72EE145 pKa = 6.44DD146 pKa = 6.22DD147 pKa = 4.6DD148 pKa = 6.78DD149 pKa = 4.88EE150 pKa = 4.89PQGDD154 pKa = 4.1WIYY157 pKa = 11.37NADD160 pKa = 3.87EE161 pKa = 4.43VEE163 pKa = 4.31SGARR167 pKa = 11.84EE168 pKa = 4.04AQMAAHH174 pKa = 7.12FDD176 pKa = 4.33SILHH180 pKa = 5.7VSPALAGGVAGQFDD194 pKa = 4.25DD195 pKa = 6.26ADD197 pKa = 4.85DD198 pKa = 4.73EE199 pKa = 5.24DD200 pKa = 6.48DD201 pKa = 3.97EE202 pKa = 5.3LLL204 pKa = 4.64
MM1 pKa = 7.3NALHH5 pKa = 6.81HH6 pKa = 7.03ADD8 pKa = 3.82CAANGAPALDD18 pKa = 3.99EE19 pKa = 5.06DD20 pKa = 4.35EE21 pKa = 5.27QILATFSRR29 pKa = 11.84TQMYY33 pKa = 8.61TNTYY37 pKa = 10.1GPDD40 pKa = 3.33EE41 pKa = 5.29DD42 pKa = 6.07DD43 pKa = 3.75NGTMEE48 pKa = 4.27HH49 pKa = 6.59KK50 pKa = 10.73GVGEE54 pKa = 4.31LYY56 pKa = 9.55VTTTRR61 pKa = 11.84IAWVFGTNWEE71 pKa = 4.13HH72 pKa = 7.55DD73 pKa = 3.42AGAVVGYY80 pKa = 9.71AWDD83 pKa = 3.5MTYY86 pKa = 11.29LSLHH90 pKa = 6.68AISRR94 pKa = 11.84DD95 pKa = 2.9ISSFRR100 pKa = 11.84FPCLYY105 pKa = 10.39CQLDD109 pKa = 3.64VEE111 pKa = 5.1DD112 pKa = 4.64EE113 pKa = 4.29VNEE116 pKa = 3.89IRR118 pKa = 11.84FVPQEE123 pKa = 4.07PEE125 pKa = 3.96KK126 pKa = 10.48EE127 pKa = 4.04LQAMFDD133 pKa = 3.77AFSASAALNPDD144 pKa = 3.72EE145 pKa = 6.44DD146 pKa = 6.22DD147 pKa = 4.6DD148 pKa = 6.78DD149 pKa = 4.88EE150 pKa = 4.89PQGDD154 pKa = 4.1WIYY157 pKa = 11.37NADD160 pKa = 3.87EE161 pKa = 4.43VEE163 pKa = 4.31SGARR167 pKa = 11.84EE168 pKa = 4.04AQMAAHH174 pKa = 7.12FDD176 pKa = 4.33SILHH180 pKa = 5.7VSPALAGGVAGQFDD194 pKa = 4.25DD195 pKa = 6.26ADD197 pKa = 4.85DD198 pKa = 4.73EE199 pKa = 5.24DD200 pKa = 6.48DD201 pKa = 3.97EE202 pKa = 5.3LLL204 pKa = 4.64
Molecular weight: 22.58 kDa
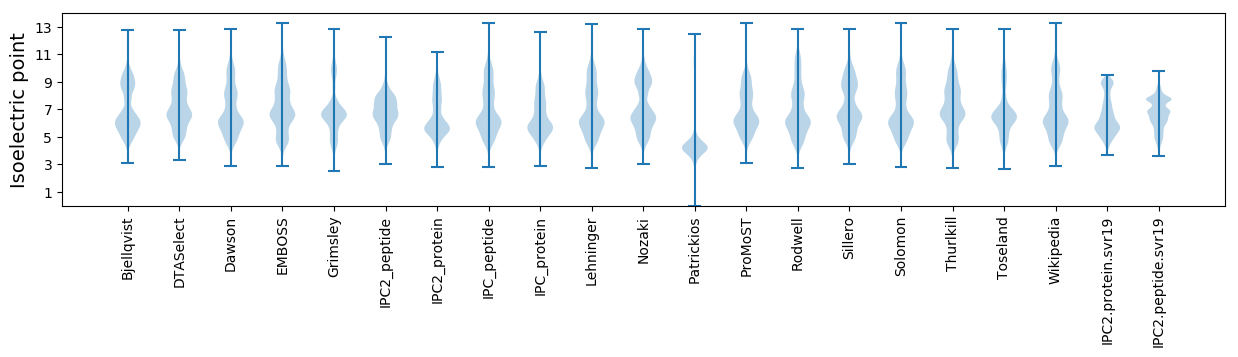
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A485LFX1|A0A485LFX1_9STRA Aste57867_20183 protein OS=Aphanomyces stellatus OX=120398 GN=Aste57867_20183 PE=3 SV=1
MM1 pKa = 7.51KK2 pKa = 10.43GLAATLLAFVLCIVLFTAQANEE24 pKa = 4.12SGLANHH30 pKa = 6.63HH31 pKa = 5.34QRR33 pKa = 11.84AVEE36 pKa = 3.97GNAMVEE42 pKa = 4.47EE43 pKa = 4.35LRR45 pKa = 11.84QLTRR49 pKa = 11.84KK50 pKa = 5.7TTAAPASSTTNTKK63 pKa = 8.6ATPTTTRR70 pKa = 11.84SSAAPAPSTTRR81 pKa = 11.84SAAPATTKK89 pKa = 9.59KK90 pKa = 6.45TTKK93 pKa = 10.25KK94 pKa = 9.44VAKK97 pKa = 9.73KK98 pKa = 9.4VAKK101 pKa = 8.88KK102 pKa = 6.8TTTKK106 pKa = 10.35TPAKK110 pKa = 9.19KK111 pKa = 6.98TTKK114 pKa = 10.25KK115 pKa = 9.56VAKK118 pKa = 9.55KK119 pKa = 10.12KK120 pKa = 10.06AVKK123 pKa = 8.83KK124 pKa = 6.1TTSAPTKK131 pKa = 10.45KK132 pKa = 8.64PTKK135 pKa = 8.78KK136 pKa = 8.59TAVKK140 pKa = 10.44KK141 pKa = 10.28DD142 pKa = 3.43VKK144 pKa = 10.5KK145 pKa = 10.83SKK147 pKa = 10.75KK148 pKa = 7.69PTKK151 pKa = 10.36KK152 pKa = 8.81PTKK155 pKa = 10.08KK156 pKa = 8.66PTKK159 pKa = 9.56KK160 pKa = 7.13TTTKK164 pKa = 10.84KK165 pKa = 10.35NVKK168 pKa = 9.56KK169 pKa = 10.64SKK171 pKa = 10.65KK172 pKa = 7.39PTKK175 pKa = 10.36KK176 pKa = 8.81PTKK179 pKa = 10.08KK180 pKa = 8.66PTKK183 pKa = 9.56KK184 pKa = 7.13TTTKK188 pKa = 10.84KK189 pKa = 10.35NVKK192 pKa = 9.56KK193 pKa = 10.64SKK195 pKa = 10.65KK196 pKa = 7.39PTKK199 pKa = 10.36KK200 pKa = 8.81PTKK203 pKa = 10.2KK204 pKa = 8.77PTKK207 pKa = 9.04KK208 pKa = 8.92TGKK211 pKa = 8.56KK212 pKa = 4.29TTKK215 pKa = 8.84KK216 pKa = 8.69TAIKK220 pKa = 9.16TADD223 pKa = 3.39KK224 pKa = 10.71KK225 pKa = 11.08AATEE229 pKa = 3.68PRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84TTRR236 pKa = 11.84KK237 pKa = 8.72PSKK240 pKa = 9.61NGKK243 pKa = 8.78KK244 pKa = 9.73AVSRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84TTRR253 pKa = 11.84KK254 pKa = 8.81PSKK257 pKa = 9.61NGKK260 pKa = 8.78KK261 pKa = 9.73AVSRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84TTRR270 pKa = 11.84KK271 pKa = 8.81PSKK274 pKa = 9.51NGKK277 pKa = 8.92KK278 pKa = 10.14AVISRR283 pKa = 11.84RR284 pKa = 11.84TTRR287 pKa = 11.84KK288 pKa = 7.64PLKK291 pKa = 8.67YY292 pKa = 9.87GKK294 pKa = 9.88KK295 pKa = 9.04AVIRR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84TTRR304 pKa = 11.84KK305 pKa = 8.83PLKK308 pKa = 9.38NGKK311 pKa = 9.17KK312 pKa = 8.94AVSRR316 pKa = 11.84RR317 pKa = 11.84RR318 pKa = 11.84TTRR321 pKa = 11.84KK322 pKa = 8.81PSKK325 pKa = 9.89NGKK328 pKa = 7.52KK329 pKa = 8.0TVIRR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84TTRR338 pKa = 11.84KK339 pKa = 8.83PLKK342 pKa = 9.38NGKK345 pKa = 9.17KK346 pKa = 8.94AVSRR350 pKa = 11.84RR351 pKa = 11.84RR352 pKa = 11.84TTRR355 pKa = 11.84KK356 pKa = 8.81PSKK359 pKa = 9.61NGKK362 pKa = 8.78KK363 pKa = 9.73AVSRR367 pKa = 11.84RR368 pKa = 11.84RR369 pKa = 11.84TTRR372 pKa = 11.84KK373 pKa = 8.81PSKK376 pKa = 9.61NGKK379 pKa = 8.78KK380 pKa = 9.73AVSRR384 pKa = 11.84RR385 pKa = 11.84RR386 pKa = 11.84TTRR389 pKa = 11.84KK390 pKa = 8.81PSKK393 pKa = 9.59NGKK396 pKa = 8.43KK397 pKa = 10.06ASIRR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84TTRR406 pKa = 11.84KK407 pKa = 9.07PSKK410 pKa = 9.57KK411 pKa = 9.56GKK413 pKa = 9.19KK414 pKa = 8.61AVSRR418 pKa = 11.84RR419 pKa = 11.84RR420 pKa = 11.84TTRR423 pKa = 11.84KK424 pKa = 8.81PSKK427 pKa = 9.61NGKK430 pKa = 8.78KK431 pKa = 9.73AVSRR435 pKa = 11.84RR436 pKa = 11.84RR437 pKa = 11.84TTRR440 pKa = 11.84KK441 pKa = 8.81PSKK444 pKa = 9.59NGKK447 pKa = 8.43KK448 pKa = 10.06ASIRR452 pKa = 11.84RR453 pKa = 11.84RR454 pKa = 11.84TTRR457 pKa = 11.84KK458 pKa = 9.07PSKK461 pKa = 9.57KK462 pKa = 9.56GKK464 pKa = 9.19KK465 pKa = 8.61AVSRR469 pKa = 11.84RR470 pKa = 11.84RR471 pKa = 11.84TTRR474 pKa = 11.84KK475 pKa = 8.81PSKK478 pKa = 9.61NGKK481 pKa = 8.78KK482 pKa = 9.73AVSRR486 pKa = 11.84RR487 pKa = 11.84RR488 pKa = 11.84TTRR491 pKa = 11.84KK492 pKa = 8.81PSKK495 pKa = 9.61NGKK498 pKa = 8.78KK499 pKa = 9.73AVSRR503 pKa = 11.84RR504 pKa = 11.84RR505 pKa = 11.84TTRR508 pKa = 11.84KK509 pKa = 8.81PSKK512 pKa = 9.88NGKK515 pKa = 9.22KK516 pKa = 9.83AVKK519 pKa = 9.75KK520 pKa = 10.39PSGKK524 pKa = 6.21TTKK527 pKa = 10.2KK528 pKa = 9.85PSKK531 pKa = 7.85TTTKK535 pKa = 10.25KK536 pKa = 10.48PVVTTSAPAAGQNATSGASAAVAVVNANN564 pKa = 3.09
MM1 pKa = 7.51KK2 pKa = 10.43GLAATLLAFVLCIVLFTAQANEE24 pKa = 4.12SGLANHH30 pKa = 6.63HH31 pKa = 5.34QRR33 pKa = 11.84AVEE36 pKa = 3.97GNAMVEE42 pKa = 4.47EE43 pKa = 4.35LRR45 pKa = 11.84QLTRR49 pKa = 11.84KK50 pKa = 5.7TTAAPASSTTNTKK63 pKa = 8.6ATPTTTRR70 pKa = 11.84SSAAPAPSTTRR81 pKa = 11.84SAAPATTKK89 pKa = 9.59KK90 pKa = 6.45TTKK93 pKa = 10.25KK94 pKa = 9.44VAKK97 pKa = 9.73KK98 pKa = 9.4VAKK101 pKa = 8.88KK102 pKa = 6.8TTTKK106 pKa = 10.35TPAKK110 pKa = 9.19KK111 pKa = 6.98TTKK114 pKa = 10.25KK115 pKa = 9.56VAKK118 pKa = 9.55KK119 pKa = 10.12KK120 pKa = 10.06AVKK123 pKa = 8.83KK124 pKa = 6.1TTSAPTKK131 pKa = 10.45KK132 pKa = 8.64PTKK135 pKa = 8.78KK136 pKa = 8.59TAVKK140 pKa = 10.44KK141 pKa = 10.28DD142 pKa = 3.43VKK144 pKa = 10.5KK145 pKa = 10.83SKK147 pKa = 10.75KK148 pKa = 7.69PTKK151 pKa = 10.36KK152 pKa = 8.81PTKK155 pKa = 10.08KK156 pKa = 8.66PTKK159 pKa = 9.56KK160 pKa = 7.13TTTKK164 pKa = 10.84KK165 pKa = 10.35NVKK168 pKa = 9.56KK169 pKa = 10.64SKK171 pKa = 10.65KK172 pKa = 7.39PTKK175 pKa = 10.36KK176 pKa = 8.81PTKK179 pKa = 10.08KK180 pKa = 8.66PTKK183 pKa = 9.56KK184 pKa = 7.13TTTKK188 pKa = 10.84KK189 pKa = 10.35NVKK192 pKa = 9.56KK193 pKa = 10.64SKK195 pKa = 10.65KK196 pKa = 7.39PTKK199 pKa = 10.36KK200 pKa = 8.81PTKK203 pKa = 10.2KK204 pKa = 8.77PTKK207 pKa = 9.04KK208 pKa = 8.92TGKK211 pKa = 8.56KK212 pKa = 4.29TTKK215 pKa = 8.84KK216 pKa = 8.69TAIKK220 pKa = 9.16TADD223 pKa = 3.39KK224 pKa = 10.71KK225 pKa = 11.08AATEE229 pKa = 3.68PRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84TTRR236 pKa = 11.84KK237 pKa = 8.72PSKK240 pKa = 9.61NGKK243 pKa = 8.78KK244 pKa = 9.73AVSRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84TTRR253 pKa = 11.84KK254 pKa = 8.81PSKK257 pKa = 9.61NGKK260 pKa = 8.78KK261 pKa = 9.73AVSRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84TTRR270 pKa = 11.84KK271 pKa = 8.81PSKK274 pKa = 9.51NGKK277 pKa = 8.92KK278 pKa = 10.14AVISRR283 pKa = 11.84RR284 pKa = 11.84TTRR287 pKa = 11.84KK288 pKa = 7.64PLKK291 pKa = 8.67YY292 pKa = 9.87GKK294 pKa = 9.88KK295 pKa = 9.04AVIRR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84TTRR304 pKa = 11.84KK305 pKa = 8.83PLKK308 pKa = 9.38NGKK311 pKa = 9.17KK312 pKa = 8.94AVSRR316 pKa = 11.84RR317 pKa = 11.84RR318 pKa = 11.84TTRR321 pKa = 11.84KK322 pKa = 8.81PSKK325 pKa = 9.89NGKK328 pKa = 7.52KK329 pKa = 8.0TVIRR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84TTRR338 pKa = 11.84KK339 pKa = 8.83PLKK342 pKa = 9.38NGKK345 pKa = 9.17KK346 pKa = 8.94AVSRR350 pKa = 11.84RR351 pKa = 11.84RR352 pKa = 11.84TTRR355 pKa = 11.84KK356 pKa = 8.81PSKK359 pKa = 9.61NGKK362 pKa = 8.78KK363 pKa = 9.73AVSRR367 pKa = 11.84RR368 pKa = 11.84RR369 pKa = 11.84TTRR372 pKa = 11.84KK373 pKa = 8.81PSKK376 pKa = 9.61NGKK379 pKa = 8.78KK380 pKa = 9.73AVSRR384 pKa = 11.84RR385 pKa = 11.84RR386 pKa = 11.84TTRR389 pKa = 11.84KK390 pKa = 8.81PSKK393 pKa = 9.59NGKK396 pKa = 8.43KK397 pKa = 10.06ASIRR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84TTRR406 pKa = 11.84KK407 pKa = 9.07PSKK410 pKa = 9.57KK411 pKa = 9.56GKK413 pKa = 9.19KK414 pKa = 8.61AVSRR418 pKa = 11.84RR419 pKa = 11.84RR420 pKa = 11.84TTRR423 pKa = 11.84KK424 pKa = 8.81PSKK427 pKa = 9.61NGKK430 pKa = 8.78KK431 pKa = 9.73AVSRR435 pKa = 11.84RR436 pKa = 11.84RR437 pKa = 11.84TTRR440 pKa = 11.84KK441 pKa = 8.81PSKK444 pKa = 9.59NGKK447 pKa = 8.43KK448 pKa = 10.06ASIRR452 pKa = 11.84RR453 pKa = 11.84RR454 pKa = 11.84TTRR457 pKa = 11.84KK458 pKa = 9.07PSKK461 pKa = 9.57KK462 pKa = 9.56GKK464 pKa = 9.19KK465 pKa = 8.61AVSRR469 pKa = 11.84RR470 pKa = 11.84RR471 pKa = 11.84TTRR474 pKa = 11.84KK475 pKa = 8.81PSKK478 pKa = 9.61NGKK481 pKa = 8.78KK482 pKa = 9.73AVSRR486 pKa = 11.84RR487 pKa = 11.84RR488 pKa = 11.84TTRR491 pKa = 11.84KK492 pKa = 8.81PSKK495 pKa = 9.61NGKK498 pKa = 8.78KK499 pKa = 9.73AVSRR503 pKa = 11.84RR504 pKa = 11.84RR505 pKa = 11.84TTRR508 pKa = 11.84KK509 pKa = 8.81PSKK512 pKa = 9.88NGKK515 pKa = 9.22KK516 pKa = 9.83AVKK519 pKa = 9.75KK520 pKa = 10.39PSGKK524 pKa = 6.21TTKK527 pKa = 10.2KK528 pKa = 9.85PSKK531 pKa = 7.85TTTKK535 pKa = 10.25KK536 pKa = 10.48PVVTTSAPAAGQNATSGASAAVAVVNANN564 pKa = 3.09
Molecular weight: 62.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9730605 |
66 |
13490 |
469.2 |
51.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.822 ± 0.021 | 1.862 ± 0.01 |
5.878 ± 0.015 | 4.892 ± 0.02 |
3.898 ± 0.012 | 5.726 ± 0.037 |
3.059 ± 0.013 | 4.223 ± 0.011 |
4.482 ± 0.019 | 9.75 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.614 ± 0.008 | 3.385 ± 0.012 |
5.337 ± 0.019 | 3.982 ± 0.011 |
5.503 ± 0.018 | 7.22 ± 0.02 |
6.618 ± 0.021 | 7.633 ± 0.016 |
1.446 ± 0.006 | 2.66 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
