
Lithobates catesbeianus (American bullfrog) (Rana catesbeiana)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amphibia; Batrachia; Anura; Neobatrachia;
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
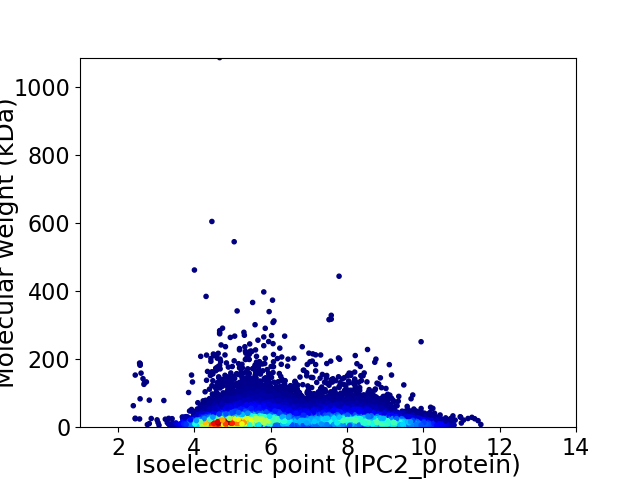
Virtual 2D-PAGE plot for 28218 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G9QE81|A0A2G9QE81_LITCT Isoform of A0A2G9QEF6 Uncharacterized protein (Fragment) OS=Lithobates catesbeianus OX=8400 GN=AB205_0084550 PE=4 SV=1
MM1 pKa = 7.6CGAEE5 pKa = 4.07QRR7 pKa = 11.84DD8 pKa = 4.62DD9 pKa = 3.96MTSHH13 pKa = 7.05CPPHH17 pKa = 7.88ISALPPSQRR26 pKa = 11.84GPLQCRR32 pKa = 11.84RR33 pKa = 11.84CGGEE37 pKa = 3.66QRR39 pKa = 11.84DD40 pKa = 4.95CIISHH45 pKa = 6.99CPPCISALLSSLNGPVLPACHH66 pKa = 5.63QCSDD70 pKa = 3.7NAHH73 pKa = 6.09LVALAGMASDD83 pKa = 3.72NTHH86 pKa = 6.68LVASDD91 pKa = 3.61VASDD95 pKa = 3.77NLQMVASDD103 pKa = 3.7VASDD107 pKa = 3.78NLQMVAGDD115 pKa = 3.76MASDD119 pKa = 3.69NLQMVVGDD127 pKa = 3.81VASDD131 pKa = 3.59NLQMVASDD139 pKa = 3.64VTSDD143 pKa = 3.48NLQMVASDD151 pKa = 3.7VASDD155 pKa = 4.04NLQMMAGDD163 pKa = 3.85MASDD167 pKa = 3.92NPHH170 pKa = 5.52QVAGDD175 pKa = 3.72VASDD179 pKa = 3.92NPHH182 pKa = 5.47QVAGDD187 pKa = 3.61VARR190 pKa = 11.84DD191 pKa = 3.71NLQMVASDD199 pKa = 3.77VQSDD203 pKa = 3.88NLKK206 pKa = 9.65MVASDD211 pKa = 3.7NLQVMASDD219 pKa = 3.85VASDD223 pKa = 3.4NLQIVEE229 pKa = 4.39GDD231 pKa = 3.61VAICKK236 pKa = 8.31WWQQ239 pKa = 2.92
MM1 pKa = 7.6CGAEE5 pKa = 4.07QRR7 pKa = 11.84DD8 pKa = 4.62DD9 pKa = 3.96MTSHH13 pKa = 7.05CPPHH17 pKa = 7.88ISALPPSQRR26 pKa = 11.84GPLQCRR32 pKa = 11.84RR33 pKa = 11.84CGGEE37 pKa = 3.66QRR39 pKa = 11.84DD40 pKa = 4.95CIISHH45 pKa = 6.99CPPCISALLSSLNGPVLPACHH66 pKa = 5.63QCSDD70 pKa = 3.7NAHH73 pKa = 6.09LVALAGMASDD83 pKa = 3.72NTHH86 pKa = 6.68LVASDD91 pKa = 3.61VASDD95 pKa = 3.77NLQMVASDD103 pKa = 3.7VASDD107 pKa = 3.78NLQMVAGDD115 pKa = 3.76MASDD119 pKa = 3.69NLQMVVGDD127 pKa = 3.81VASDD131 pKa = 3.59NLQMVASDD139 pKa = 3.64VTSDD143 pKa = 3.48NLQMVASDD151 pKa = 3.7VASDD155 pKa = 4.04NLQMMAGDD163 pKa = 3.85MASDD167 pKa = 3.92NPHH170 pKa = 5.52QVAGDD175 pKa = 3.72VASDD179 pKa = 3.92NPHH182 pKa = 5.47QVAGDD187 pKa = 3.61VARR190 pKa = 11.84DD191 pKa = 3.71NLQMVASDD199 pKa = 3.77VQSDD203 pKa = 3.88NLKK206 pKa = 9.65MVASDD211 pKa = 3.7NLQVMASDD219 pKa = 3.85VASDD223 pKa = 3.4NLQIVEE229 pKa = 4.39GDD231 pKa = 3.61VAICKK236 pKa = 8.31WWQQ239 pKa = 2.92
Molecular weight: 25.11 kDa
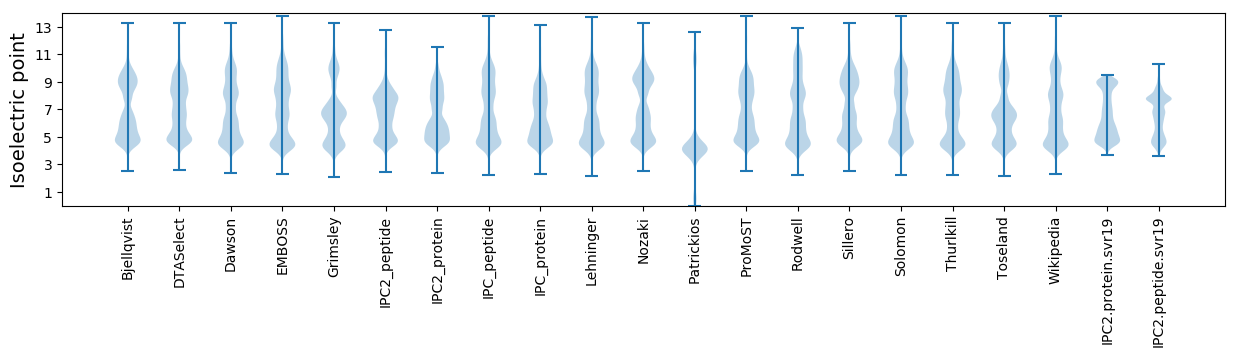
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G9QM72|A0A2G9QM72_LITCT SERPIN domain-containing protein OS=Lithobates catesbeianus OX=8400 GN=AB205_0132410 PE=3 SV=1
MM1 pKa = 7.27KK2 pKa = 10.52VSIRR6 pKa = 11.84GTRR9 pKa = 11.84RR10 pKa = 11.84IGVRR14 pKa = 11.84VTGRR18 pKa = 11.84VGVKK22 pKa = 9.76VIISGTRR29 pKa = 11.84RR30 pKa = 11.84VVIRR34 pKa = 11.84VTGKK38 pKa = 10.38ASVKK42 pKa = 9.86VSIRR46 pKa = 11.84GTRR49 pKa = 11.84RR50 pKa = 11.84AGVRR54 pKa = 11.84ITGRR58 pKa = 11.84VGVKK62 pKa = 9.54ISNRR66 pKa = 11.84GTRR69 pKa = 11.84RR70 pKa = 11.84VGFRR74 pKa = 11.84VAGRR78 pKa = 11.84VGVKK82 pKa = 10.14ARR84 pKa = 11.84IRR86 pKa = 11.84GTRR89 pKa = 11.84RR90 pKa = 11.84VGIRR94 pKa = 11.84VTGRR98 pKa = 11.84ASVKK102 pKa = 9.78VSIRR106 pKa = 11.84GTRR109 pKa = 11.84RR110 pKa = 11.84AGVRR114 pKa = 11.84VTGRR118 pKa = 11.84VSVKK122 pKa = 9.83VSNRR126 pKa = 11.84GAGRR130 pKa = 11.84VGVRR134 pKa = 11.84VTSRR138 pKa = 11.84VGVKK142 pKa = 9.55VSNRR146 pKa = 11.84GTRR149 pKa = 11.84RR150 pKa = 11.84VGFRR154 pKa = 11.84VAGRR158 pKa = 11.84VGVKK162 pKa = 9.58VRR164 pKa = 11.84IRR166 pKa = 11.84GPRR169 pKa = 11.84RR170 pKa = 11.84VGIRR174 pKa = 11.84VTGRR178 pKa = 11.84ASVKK182 pKa = 9.69VSNRR186 pKa = 11.84GTRR189 pKa = 11.84RR190 pKa = 11.84VGVRR194 pKa = 11.84VSDD197 pKa = 3.35GGTRR201 pKa = 11.84RR202 pKa = 11.84DD203 pKa = 3.45GFRR206 pKa = 11.84VAGKK210 pKa = 10.42VDD212 pKa = 3.44VKK214 pKa = 10.93LSIRR218 pKa = 11.84GTRR221 pKa = 11.84RR222 pKa = 11.84VGFRR226 pKa = 11.84VTGRR230 pKa = 3.29
MM1 pKa = 7.27KK2 pKa = 10.52VSIRR6 pKa = 11.84GTRR9 pKa = 11.84RR10 pKa = 11.84IGVRR14 pKa = 11.84VTGRR18 pKa = 11.84VGVKK22 pKa = 9.76VIISGTRR29 pKa = 11.84RR30 pKa = 11.84VVIRR34 pKa = 11.84VTGKK38 pKa = 10.38ASVKK42 pKa = 9.86VSIRR46 pKa = 11.84GTRR49 pKa = 11.84RR50 pKa = 11.84AGVRR54 pKa = 11.84ITGRR58 pKa = 11.84VGVKK62 pKa = 9.54ISNRR66 pKa = 11.84GTRR69 pKa = 11.84RR70 pKa = 11.84VGFRR74 pKa = 11.84VAGRR78 pKa = 11.84VGVKK82 pKa = 10.14ARR84 pKa = 11.84IRR86 pKa = 11.84GTRR89 pKa = 11.84RR90 pKa = 11.84VGIRR94 pKa = 11.84VTGRR98 pKa = 11.84ASVKK102 pKa = 9.78VSIRR106 pKa = 11.84GTRR109 pKa = 11.84RR110 pKa = 11.84AGVRR114 pKa = 11.84VTGRR118 pKa = 11.84VSVKK122 pKa = 9.83VSNRR126 pKa = 11.84GAGRR130 pKa = 11.84VGVRR134 pKa = 11.84VTSRR138 pKa = 11.84VGVKK142 pKa = 9.55VSNRR146 pKa = 11.84GTRR149 pKa = 11.84RR150 pKa = 11.84VGFRR154 pKa = 11.84VAGRR158 pKa = 11.84VGVKK162 pKa = 9.58VRR164 pKa = 11.84IRR166 pKa = 11.84GPRR169 pKa = 11.84RR170 pKa = 11.84VGIRR174 pKa = 11.84VTGRR178 pKa = 11.84ASVKK182 pKa = 9.69VSNRR186 pKa = 11.84GTRR189 pKa = 11.84RR190 pKa = 11.84VGVRR194 pKa = 11.84VSDD197 pKa = 3.35GGTRR201 pKa = 11.84RR202 pKa = 11.84DD203 pKa = 3.45GFRR206 pKa = 11.84VAGKK210 pKa = 10.42VDD212 pKa = 3.44VKK214 pKa = 10.93LSIRR218 pKa = 11.84GTRR221 pKa = 11.84RR222 pKa = 11.84VGFRR226 pKa = 11.84VTGRR230 pKa = 3.29
Molecular weight: 24.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7230904 |
24 |
9797 |
256.3 |
28.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.498 ± 0.016 | 2.471 ± 0.015 |
5.083 ± 0.015 | 7.21 ± 0.031 |
3.577 ± 0.014 | 5.973 ± 0.024 |
2.894 ± 0.012 | 5.161 ± 0.015 |
5.886 ± 0.021 | 8.805 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.457 ± 0.01 | 4.056 ± 0.015 |
5.968 ± 0.025 | 4.727 ± 0.016 |
5.311 ± 0.017 | 8.834 ± 0.025 |
5.905 ± 0.025 | 6.179 ± 0.028 |
1.055 ± 0.006 | 2.943 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
