
Thiohalocapsa sp. PB-PSB1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Thiohalocapsa; unclassified Thiohalocapsa
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
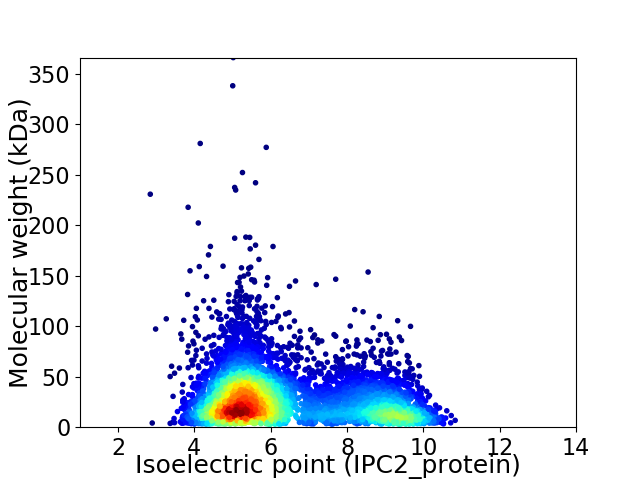
Virtual 2D-PAGE plot for 6200 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V4JJW4|V4JJW4_9GAMM Uncharacterized protein (Fragment) OS=Thiohalocapsa sp. PB-PSB1 OX=1385625 GN=N838_30920 PE=4 SV=1
MM1 pKa = 5.93THH3 pKa = 7.01KK4 pKa = 9.97RR5 pKa = 11.84TAWWNRR11 pKa = 11.84LALSAIVGLLAAIGTLTAAPSRR33 pKa = 11.84AEE35 pKa = 4.14TIAYY39 pKa = 9.61GYY41 pKa = 10.97DD42 pKa = 3.03PGNRR46 pKa = 11.84LEE48 pKa = 4.16SVEE51 pKa = 4.15YY52 pKa = 10.7DD53 pKa = 3.2DD54 pKa = 4.96GQSIRR59 pKa = 11.84YY60 pKa = 9.58VYY62 pKa = 11.01DD63 pKa = 3.12NLGNQLVRR71 pKa = 11.84LTLAAPEE78 pKa = 4.39ANNPPGTVTPGVADD92 pKa = 3.91GTEE95 pKa = 3.9LTYY98 pKa = 10.29TAVTLDD104 pKa = 3.19WSAAVDD110 pKa = 4.01PDD112 pKa = 3.64AGDD115 pKa = 3.21AVVYY119 pKa = 9.5YY120 pKa = 9.77LHH122 pKa = 7.27LGTEE126 pKa = 4.36ADD128 pKa = 3.78PPLVYY133 pKa = 10.68SGWEE137 pKa = 3.56RR138 pKa = 11.84SYY140 pKa = 10.94TPPYY144 pKa = 10.25SLQPLTTYY152 pKa = 9.75FWSVVARR159 pKa = 11.84DD160 pKa = 3.77SQGAEE165 pKa = 4.1SVSGPWTFITGNEE178 pKa = 4.25PPTPIIDD185 pKa = 4.08ASPTEE190 pKa = 4.66AIVPYY195 pKa = 9.17TANLTDD201 pKa = 4.3ASTSTDD207 pKa = 3.24DD208 pKa = 5.62AIASRR213 pKa = 11.84AWDD216 pKa = 4.07FLCDD220 pKa = 3.85GSIDD224 pKa = 3.81ATTSTASYY232 pKa = 9.19YY233 pKa = 10.89VSAAGDD239 pKa = 3.8YY240 pKa = 7.74EE241 pKa = 3.98ICLAVTDD248 pKa = 4.12EE249 pKa = 4.55AGATEE254 pKa = 4.38STSVTLYY261 pKa = 11.09GRR263 pKa = 11.84IDD265 pKa = 3.53TDD267 pKa = 3.4GDD269 pKa = 4.21GVFDD273 pKa = 5.04IADD276 pKa = 3.9NCPADD281 pKa = 4.37ANADD285 pKa = 3.77QYY287 pKa = 12.23NLDD290 pKa = 4.16GDD292 pKa = 4.45DD293 pKa = 5.34LGDD296 pKa = 3.59VCDD299 pKa = 5.55DD300 pKa = 4.36DD301 pKa = 6.37RR302 pKa = 11.84DD303 pKa = 4.01GDD305 pKa = 4.31GVTNAADD312 pKa = 4.29PFPDD316 pKa = 3.82DD317 pKa = 3.81ARR319 pKa = 11.84YY320 pKa = 7.47TADD323 pKa = 3.73SDD325 pKa = 3.88GDD327 pKa = 4.22GIGDD331 pKa = 3.24SWEE334 pKa = 3.75IAEE337 pKa = 4.8FGDD340 pKa = 6.22LITADD345 pKa = 4.12DD346 pKa = 4.0TTDD349 pKa = 3.62FDD351 pKa = 5.44GDD353 pKa = 3.72GSSDD357 pKa = 3.61LDD359 pKa = 3.53EE360 pKa = 4.87FRR362 pKa = 11.84YY363 pKa = 10.78DD364 pKa = 4.21SDD366 pKa = 3.63PQYY369 pKa = 11.55APPFVDD375 pKa = 3.76TAPIAAGGDD384 pKa = 3.19HH385 pKa = 6.92SLAQRR390 pKa = 11.84TDD392 pKa = 2.59GRR394 pKa = 11.84LYY396 pKa = 10.04AWGYY400 pKa = 9.58NGNGQLGDD408 pKa = 3.47GSYY411 pKa = 11.06TNRR414 pKa = 11.84NYY416 pKa = 10.41PAFVRR421 pKa = 11.84DD422 pKa = 3.47DD423 pKa = 3.49TGAFVYY429 pKa = 10.22DD430 pKa = 3.74IQDD433 pKa = 3.01IAAGGDD439 pKa = 3.14HH440 pKa = 6.61SLAVLGDD447 pKa = 3.61GSLISWGYY455 pKa = 9.77NGYY458 pKa = 9.23GQLGDD463 pKa = 3.62GTNTRR468 pKa = 11.84RR469 pKa = 11.84ADD471 pKa = 3.59PVAVIDD477 pKa = 4.75GYY479 pKa = 11.46GDD481 pKa = 3.67PVIDD485 pKa = 4.13AVSVAAGANHH495 pKa = 6.58SLALLDD501 pKa = 5.54DD502 pKa = 4.75GSLLAWGYY510 pKa = 10.91NGYY513 pKa = 9.24GQLGDD518 pKa = 3.55GTTTSKK524 pKa = 9.36STPVRR529 pKa = 11.84VRR531 pKa = 11.84DD532 pKa = 3.75AVGDD536 pKa = 3.83TLTGIIAVAAGQNFSLALRR555 pKa = 11.84SDD557 pKa = 3.53GTVWAWGRR565 pKa = 11.84NYY567 pKa = 10.07QGQLGDD573 pKa = 3.85GTNSNSASAQRR584 pKa = 11.84VLDD587 pKa = 3.79ARR589 pKa = 11.84GMPINGVVAVAAGFRR604 pKa = 11.84HH605 pKa = 5.91GLALSEE611 pKa = 4.84AGTAVAWGDD620 pKa = 3.21NDD622 pKa = 3.67YY623 pKa = 11.45GQLGDD628 pKa = 4.05GFTVDD633 pKa = 3.04RR634 pKa = 11.84RR635 pKa = 11.84VPVQVIDD642 pKa = 4.02AFGTPVDD649 pKa = 4.11GMSALAAGEE658 pKa = 4.09WHH660 pKa = 6.52SLALASGGDD669 pKa = 3.51VLAWGRR675 pKa = 11.84NNLGQLGEE683 pKa = 4.43GTLSDD688 pKa = 3.7SSIPTSLGLTGITGIDD704 pKa = 3.28AGSEE708 pKa = 3.77HH709 pKa = 6.75SLAVTDD715 pKa = 5.35DD716 pKa = 3.54LTAFAWGRR724 pKa = 11.84NALFQLGDD732 pKa = 3.35GTATDD737 pKa = 3.34RR738 pKa = 11.84RR739 pKa = 11.84SPVVVIDD746 pKa = 4.06NNLAPIFPVGKK757 pKa = 9.65PSTDD761 pKa = 3.32PAADD765 pKa = 3.41SDD767 pKa = 4.47GDD769 pKa = 4.19GEE771 pKa = 4.63PDD773 pKa = 3.34STDD776 pKa = 2.99AFPYY780 pKa = 10.48DD781 pKa = 3.09PAYY784 pKa = 10.87RR785 pKa = 11.84LDD787 pKa = 3.85ADD789 pKa = 3.93GDD791 pKa = 4.34GIADD795 pKa = 3.54EE796 pKa = 4.54WEE798 pKa = 3.97LANFGDD804 pKa = 5.35LLTAGIGTDD813 pKa = 3.58YY814 pKa = 11.47DD815 pKa = 4.17GDD817 pKa = 3.92GLLDD821 pKa = 3.65SEE823 pKa = 4.55EE824 pKa = 4.53FEE826 pKa = 4.8HH827 pKa = 8.0GSDD830 pKa = 3.09ATVAPEE836 pKa = 3.79MVEE839 pKa = 4.63DD840 pKa = 4.46VVATGANHH848 pKa = 6.1TLALRR853 pKa = 11.84TDD855 pKa = 3.45GRR857 pKa = 11.84VLAWGYY863 pKa = 10.87NGYY866 pKa = 9.22GQLGNGTTQNQSSVSYY882 pKa = 8.34THH884 pKa = 6.94
MM1 pKa = 5.93THH3 pKa = 7.01KK4 pKa = 9.97RR5 pKa = 11.84TAWWNRR11 pKa = 11.84LALSAIVGLLAAIGTLTAAPSRR33 pKa = 11.84AEE35 pKa = 4.14TIAYY39 pKa = 9.61GYY41 pKa = 10.97DD42 pKa = 3.03PGNRR46 pKa = 11.84LEE48 pKa = 4.16SVEE51 pKa = 4.15YY52 pKa = 10.7DD53 pKa = 3.2DD54 pKa = 4.96GQSIRR59 pKa = 11.84YY60 pKa = 9.58VYY62 pKa = 11.01DD63 pKa = 3.12NLGNQLVRR71 pKa = 11.84LTLAAPEE78 pKa = 4.39ANNPPGTVTPGVADD92 pKa = 3.91GTEE95 pKa = 3.9LTYY98 pKa = 10.29TAVTLDD104 pKa = 3.19WSAAVDD110 pKa = 4.01PDD112 pKa = 3.64AGDD115 pKa = 3.21AVVYY119 pKa = 9.5YY120 pKa = 9.77LHH122 pKa = 7.27LGTEE126 pKa = 4.36ADD128 pKa = 3.78PPLVYY133 pKa = 10.68SGWEE137 pKa = 3.56RR138 pKa = 11.84SYY140 pKa = 10.94TPPYY144 pKa = 10.25SLQPLTTYY152 pKa = 9.75FWSVVARR159 pKa = 11.84DD160 pKa = 3.77SQGAEE165 pKa = 4.1SVSGPWTFITGNEE178 pKa = 4.25PPTPIIDD185 pKa = 4.08ASPTEE190 pKa = 4.66AIVPYY195 pKa = 9.17TANLTDD201 pKa = 4.3ASTSTDD207 pKa = 3.24DD208 pKa = 5.62AIASRR213 pKa = 11.84AWDD216 pKa = 4.07FLCDD220 pKa = 3.85GSIDD224 pKa = 3.81ATTSTASYY232 pKa = 9.19YY233 pKa = 10.89VSAAGDD239 pKa = 3.8YY240 pKa = 7.74EE241 pKa = 3.98ICLAVTDD248 pKa = 4.12EE249 pKa = 4.55AGATEE254 pKa = 4.38STSVTLYY261 pKa = 11.09GRR263 pKa = 11.84IDD265 pKa = 3.53TDD267 pKa = 3.4GDD269 pKa = 4.21GVFDD273 pKa = 5.04IADD276 pKa = 3.9NCPADD281 pKa = 4.37ANADD285 pKa = 3.77QYY287 pKa = 12.23NLDD290 pKa = 4.16GDD292 pKa = 4.45DD293 pKa = 5.34LGDD296 pKa = 3.59VCDD299 pKa = 5.55DD300 pKa = 4.36DD301 pKa = 6.37RR302 pKa = 11.84DD303 pKa = 4.01GDD305 pKa = 4.31GVTNAADD312 pKa = 4.29PFPDD316 pKa = 3.82DD317 pKa = 3.81ARR319 pKa = 11.84YY320 pKa = 7.47TADD323 pKa = 3.73SDD325 pKa = 3.88GDD327 pKa = 4.22GIGDD331 pKa = 3.24SWEE334 pKa = 3.75IAEE337 pKa = 4.8FGDD340 pKa = 6.22LITADD345 pKa = 4.12DD346 pKa = 4.0TTDD349 pKa = 3.62FDD351 pKa = 5.44GDD353 pKa = 3.72GSSDD357 pKa = 3.61LDD359 pKa = 3.53EE360 pKa = 4.87FRR362 pKa = 11.84YY363 pKa = 10.78DD364 pKa = 4.21SDD366 pKa = 3.63PQYY369 pKa = 11.55APPFVDD375 pKa = 3.76TAPIAAGGDD384 pKa = 3.19HH385 pKa = 6.92SLAQRR390 pKa = 11.84TDD392 pKa = 2.59GRR394 pKa = 11.84LYY396 pKa = 10.04AWGYY400 pKa = 9.58NGNGQLGDD408 pKa = 3.47GSYY411 pKa = 11.06TNRR414 pKa = 11.84NYY416 pKa = 10.41PAFVRR421 pKa = 11.84DD422 pKa = 3.47DD423 pKa = 3.49TGAFVYY429 pKa = 10.22DD430 pKa = 3.74IQDD433 pKa = 3.01IAAGGDD439 pKa = 3.14HH440 pKa = 6.61SLAVLGDD447 pKa = 3.61GSLISWGYY455 pKa = 9.77NGYY458 pKa = 9.23GQLGDD463 pKa = 3.62GTNTRR468 pKa = 11.84RR469 pKa = 11.84ADD471 pKa = 3.59PVAVIDD477 pKa = 4.75GYY479 pKa = 11.46GDD481 pKa = 3.67PVIDD485 pKa = 4.13AVSVAAGANHH495 pKa = 6.58SLALLDD501 pKa = 5.54DD502 pKa = 4.75GSLLAWGYY510 pKa = 10.91NGYY513 pKa = 9.24GQLGDD518 pKa = 3.55GTTTSKK524 pKa = 9.36STPVRR529 pKa = 11.84VRR531 pKa = 11.84DD532 pKa = 3.75AVGDD536 pKa = 3.83TLTGIIAVAAGQNFSLALRR555 pKa = 11.84SDD557 pKa = 3.53GTVWAWGRR565 pKa = 11.84NYY567 pKa = 10.07QGQLGDD573 pKa = 3.85GTNSNSASAQRR584 pKa = 11.84VLDD587 pKa = 3.79ARR589 pKa = 11.84GMPINGVVAVAAGFRR604 pKa = 11.84HH605 pKa = 5.91GLALSEE611 pKa = 4.84AGTAVAWGDD620 pKa = 3.21NDD622 pKa = 3.67YY623 pKa = 11.45GQLGDD628 pKa = 4.05GFTVDD633 pKa = 3.04RR634 pKa = 11.84RR635 pKa = 11.84VPVQVIDD642 pKa = 4.02AFGTPVDD649 pKa = 4.11GMSALAAGEE658 pKa = 4.09WHH660 pKa = 6.52SLALASGGDD669 pKa = 3.51VLAWGRR675 pKa = 11.84NNLGQLGEE683 pKa = 4.43GTLSDD688 pKa = 3.7SSIPTSLGLTGITGIDD704 pKa = 3.28AGSEE708 pKa = 3.77HH709 pKa = 6.75SLAVTDD715 pKa = 5.35DD716 pKa = 3.54LTAFAWGRR724 pKa = 11.84NALFQLGDD732 pKa = 3.35GTATDD737 pKa = 3.34RR738 pKa = 11.84RR739 pKa = 11.84SPVVVIDD746 pKa = 4.06NNLAPIFPVGKK757 pKa = 9.65PSTDD761 pKa = 3.32PAADD765 pKa = 3.41SDD767 pKa = 4.47GDD769 pKa = 4.19GEE771 pKa = 4.63PDD773 pKa = 3.34STDD776 pKa = 2.99AFPYY780 pKa = 10.48DD781 pKa = 3.09PAYY784 pKa = 10.87RR785 pKa = 11.84LDD787 pKa = 3.85ADD789 pKa = 3.93GDD791 pKa = 4.34GIADD795 pKa = 3.54EE796 pKa = 4.54WEE798 pKa = 3.97LANFGDD804 pKa = 5.35LLTAGIGTDD813 pKa = 3.58YY814 pKa = 11.47DD815 pKa = 4.17GDD817 pKa = 3.92GLLDD821 pKa = 3.65SEE823 pKa = 4.55EE824 pKa = 4.53FEE826 pKa = 4.8HH827 pKa = 8.0GSDD830 pKa = 3.09ATVAPEE836 pKa = 3.79MVEE839 pKa = 4.63DD840 pKa = 4.46VVATGANHH848 pKa = 6.1TLALRR853 pKa = 11.84TDD855 pKa = 3.45GRR857 pKa = 11.84VLAWGYY863 pKa = 10.87NGYY866 pKa = 9.22GQLGNGTTQNQSSVSYY882 pKa = 8.34THH884 pKa = 6.94
Molecular weight: 92.37 kDa
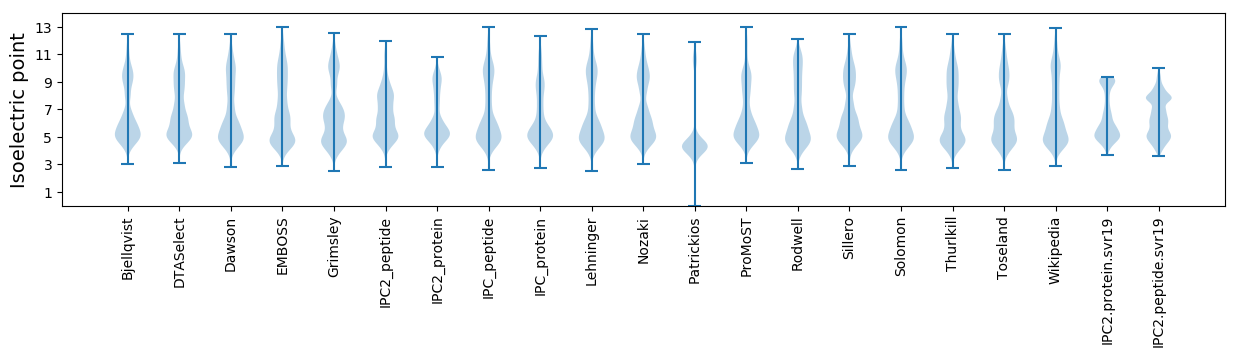
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V4JDW3|V4JDW3_9GAMM Oxidoreductase OS=Thiohalocapsa sp. PB-PSB1 OX=1385625 GN=N838_02070 PE=4 SV=1
KKK2 pKa = 9.53WRR4 pKa = 11.84RR5 pKa = 11.84ALYYY9 pKa = 10.1QLRR12 pKa = 11.84RR13 pKa = 11.84HHH15 pKa = 6.08GGVFRR20 pKa = 11.84EEE22 pKa = 4.33ARR24 pKa = 11.84HHH26 pKa = 5.39KK27 pKa = 10.49DD28 pKa = 3.2RR29 pKa = 11.84IEEE32 pKa = 3.94EE33 pKa = 4.17HHH35 pKa = 5.89MPDDD39 pKa = 3.4HH40 pKa = 5.82HHH42 pKa = 5.8PISIPPKKK50 pKa = 8.98DD51 pKa = 3.59VSQVVGYYY59 pKa = 10.2RR60 pKa = 11.84GKKK63 pKa = 10.03AIHHH67 pKa = 6.48ARR69 pKa = 11.84VYYY72 pKa = 10.38EEE74 pKa = 4.06KKK76 pKa = 9.25NFAGQHHH83 pKa = 4.7RR84 pKa = 11.84ARR86 pKa = 11.84GCFVSTVGRR95 pKa = 11.84DDD97 pKa = 2.94EE98 pKa = 4.21TRR100 pKa = 11.84SEEE103 pKa = 4.42TSSTSNVKKK112 pKa = 10.19KKK114 pKa = 10.92DD115 pKa = 3.86IN
KKK2 pKa = 9.53WRR4 pKa = 11.84RR5 pKa = 11.84ALYYY9 pKa = 10.1QLRR12 pKa = 11.84RR13 pKa = 11.84HHH15 pKa = 6.08GGVFRR20 pKa = 11.84EEE22 pKa = 4.33ARR24 pKa = 11.84HHH26 pKa = 5.39KK27 pKa = 10.49DD28 pKa = 3.2RR29 pKa = 11.84IEEE32 pKa = 3.94EE33 pKa = 4.17HHH35 pKa = 5.89MPDDD39 pKa = 3.4HH40 pKa = 5.82HHH42 pKa = 5.8PISIPPKKK50 pKa = 8.98DD51 pKa = 3.59VSQVVGYYY59 pKa = 10.2RR60 pKa = 11.84GKKK63 pKa = 10.03AIHHH67 pKa = 6.48ARR69 pKa = 11.84VYYY72 pKa = 10.38EEE74 pKa = 4.06KKK76 pKa = 9.25NFAGQHHH83 pKa = 4.7RR84 pKa = 11.84ARR86 pKa = 11.84GCFVSTVGRR95 pKa = 11.84DDD97 pKa = 2.94EE98 pKa = 4.21TRR100 pKa = 11.84SEEE103 pKa = 4.42TSSTSNVKKK112 pKa = 10.19KKK114 pKa = 10.92DD115 pKa = 3.86IN
Molecular weight: 13.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1699814 |
29 |
3305 |
274.2 |
30.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.996 ± 0.041 | 1.105 ± 0.014 |
6.178 ± 0.029 | 5.759 ± 0.033 |
3.583 ± 0.02 | 7.706 ± 0.031 |
2.323 ± 0.016 | 5.139 ± 0.024 |
2.863 ± 0.024 | 11.208 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.269 ± 0.016 | 2.888 ± 0.022 |
5.213 ± 0.026 | 4.109 ± 0.024 |
7.521 ± 0.04 | 5.654 ± 0.028 |
4.935 ± 0.025 | 6.576 ± 0.03 |
1.491 ± 0.016 | 2.483 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
