
Chromatium okenii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Chromatium
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
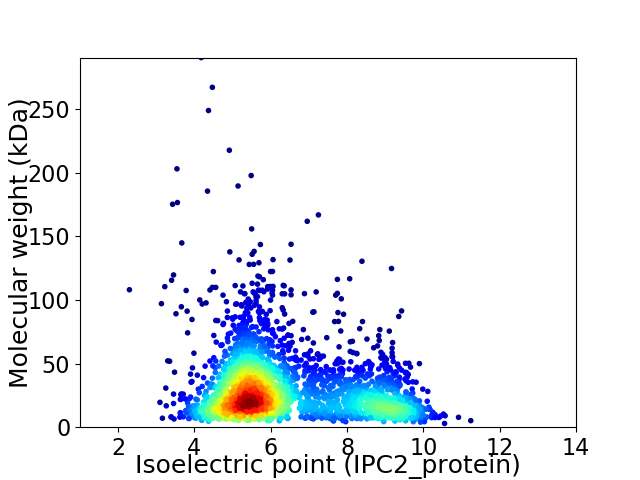
Virtual 2D-PAGE plot for 2811 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S7XNB3|A0A2S7XNB3_9GAMM RimK family alpha-L-glutamate ligase OS=Chromatium okenii OX=61644 GN=CXB77_13135 PE=4 SV=1
MM1 pKa = 6.83FASRR5 pKa = 11.84AHH7 pKa = 6.18AVLITDD13 pKa = 4.73AQLQDD18 pKa = 4.44LDD20 pKa = 3.85QLLAGLDD27 pKa = 3.5ADD29 pKa = 3.99VEE31 pKa = 4.38PWLVASNQDD40 pKa = 3.28AMPLIFKK47 pKa = 10.64ALAHH51 pKa = 6.95PDD53 pKa = 3.39LTQLHH58 pKa = 6.79LLAHH62 pKa = 6.69GAPGEE67 pKa = 3.96IRR69 pKa = 11.84LGEE72 pKa = 4.17RR73 pKa = 11.84ALTAADD79 pKa = 3.6FRR81 pKa = 11.84QYY83 pKa = 11.5LNGAAMRR90 pKa = 11.84DD91 pKa = 3.42LSINFWSCHH100 pKa = 4.94TGAGEE105 pKa = 3.87MGTEE109 pKa = 4.24FTHH112 pKa = 6.96AVAAATGARR121 pKa = 11.84VLAAEE126 pKa = 4.31GLIGAANKK134 pKa = 9.52QGSWEE139 pKa = 4.23LNGITAPFSEE149 pKa = 4.82TARR152 pKa = 11.84AGYY155 pKa = 11.06ANVLAVRR162 pKa = 11.84DD163 pKa = 3.86RR164 pKa = 11.84FEE166 pKa = 6.41DD167 pKa = 3.39NDD169 pKa = 3.98TAATATNLGTITSTRR184 pKa = 11.84TEE186 pKa = 3.99TGLSIEE192 pKa = 4.66TNDD195 pKa = 3.85PDD197 pKa = 3.39WFQFTLGSTGTSASNVAIDD216 pKa = 3.96FSNALGDD223 pKa = 4.36LDD225 pKa = 4.21IFLYY229 pKa = 10.76DD230 pKa = 3.32AAGNALQGTYY240 pKa = 10.42GSNDD244 pKa = 3.22HH245 pKa = 6.04EE246 pKa = 4.61QISLSGLAAGSYY258 pKa = 9.66LIKK261 pKa = 10.85VRR263 pKa = 11.84DD264 pKa = 3.6WTSLDD269 pKa = 3.22GGEE272 pKa = 4.28NPNYY276 pKa = 9.45TLTLIGSGEE285 pKa = 4.03VSPTVSYY292 pKa = 7.64PTLDD296 pKa = 3.51YY297 pKa = 10.98MPDD300 pKa = 3.37LTSSQMSGARR310 pKa = 11.84SLTFAPYY317 pKa = 10.25TSTSWDD323 pKa = 3.59SKK325 pKa = 9.78AGAYY329 pKa = 10.06GSEE332 pKa = 4.46SGASSHH338 pKa = 7.05SIYY341 pKa = 10.43TITATAGQVYY351 pKa = 10.44DD352 pKa = 5.02LYY354 pKa = 11.03SVSYY358 pKa = 9.78FDD360 pKa = 4.79PYY362 pKa = 11.5SLTVYY367 pKa = 10.59DD368 pKa = 4.15SLGNAIVTNAEE379 pKa = 3.69SDD381 pKa = 3.82DD382 pKa = 3.94PADD385 pKa = 3.8YY386 pKa = 11.43NLGDD390 pKa = 3.77AYY392 pKa = 11.47YY393 pKa = 10.88GVDD396 pKa = 3.93WLNDD400 pKa = 2.87WTAPYY405 pKa = 10.47SGTYY409 pKa = 9.58YY410 pKa = 10.93VDD412 pKa = 4.35ASWHH416 pKa = 5.08QGSYY420 pKa = 7.75FTFYY424 pKa = 11.62DD425 pKa = 3.9LMIYY429 pKa = 9.93KK430 pKa = 10.29NKK432 pKa = 9.06STTVLPTLSLSSLSPSSVSEE452 pKa = 4.22GNSGSTSMMVNVTRR466 pKa = 11.84TGDD469 pKa = 3.45LSSTSSASWSVVGAAATASDD489 pKa = 4.27FLSGVLPTGTVNFAAGQASATATINIAGDD518 pKa = 3.46TTIEE522 pKa = 3.84SDD524 pKa = 3.76EE525 pKa = 4.39NFTVTLSNPVSATLDD540 pKa = 3.16ISSATGTIVNDD551 pKa = 3.98DD552 pKa = 3.61VSPPVPYY559 pKa = 8.19PTLDD563 pKa = 3.75YY564 pKa = 11.1IPDD567 pKa = 3.97LKK569 pKa = 10.8SSQMYY574 pKa = 9.81EE575 pKa = 3.75ADD577 pKa = 4.08LLAFLPVTSSVYY589 pKa = 10.47YY590 pKa = 10.92DD591 pKa = 3.43MEE593 pKa = 6.61AYY595 pKa = 10.15DD596 pKa = 4.9YY597 pKa = 11.29GLTSNASLHH606 pKa = 6.44SLYY609 pKa = 10.62QFNATTGVLYY619 pKa = 10.32DD620 pKa = 4.35IYY622 pKa = 10.65SISYY626 pKa = 10.41LDD628 pKa = 3.63PTEE631 pKa = 3.93LTIYY635 pKa = 10.41DD636 pKa = 3.6QFGNAIAINNEE647 pKa = 3.52SDD649 pKa = 4.64DD650 pKa = 4.42PDD652 pKa = 3.7NSTLSWVDD660 pKa = 3.36DD661 pKa = 3.99GGSYY665 pKa = 10.79SFDD668 pKa = 5.43AIWEE672 pKa = 3.95WSAPYY677 pKa = 10.47SGTFYY682 pKa = 11.24VDD684 pKa = 3.34SGWNQGSFFTYY695 pKa = 9.14WDD697 pKa = 3.25VSVYY701 pKa = 10.25RR702 pKa = 11.84NKK704 pKa = 9.91STTPSLPTLSLSSLTPTSISEE725 pKa = 4.32GNSGYY730 pKa = 9.0TPITFTVNRR739 pKa = 11.84DD740 pKa = 2.9GDD742 pKa = 3.73LSGTSRR748 pKa = 11.84VFFAVTGTGTNPVNAADD765 pKa = 4.08FRR767 pKa = 11.84DD768 pKa = 4.06GVLPSDD774 pKa = 4.47WIDD777 pKa = 3.57FDD779 pKa = 4.13SGAVSATITSYY790 pKa = 11.3VVAGDD795 pKa = 3.59TTPEE799 pKa = 3.95PNEE802 pKa = 4.03NFTVTLSSPTGATLGTSTSVIGTITNDD829 pKa = 3.6DD830 pKa = 4.35SLPLPTISLSPLSPSIITEE849 pKa = 3.91GHH851 pKa = 5.52SGYY854 pKa = 8.97TPVNIVVTRR863 pKa = 11.84GGDD866 pKa = 3.41LSSTSSATWTVVGTGVAPVTADD888 pKa = 3.45DD889 pKa = 4.54FGGFLPSGIISLAAGVATATVSINILGDD917 pKa = 3.39TTVEE921 pKa = 4.18SNEE924 pKa = 4.54DD925 pKa = 3.49FTVMLSNPSGATLIPNTVTGTITNDD950 pKa = 3.32DD951 pKa = 4.28VILPTLSLASLTPSIIMEE969 pKa = 4.48GNSGFTPMTVTITRR983 pKa = 11.84SGDD986 pKa = 3.4LSGTSAANWSVIGAGATADD1005 pKa = 3.81DD1006 pKa = 4.95FIGGVLPSGTISFPAWQATTTATINIFGDD1035 pKa = 3.62TLVEE1039 pKa = 4.38SNEE1042 pKa = 3.95NFSVVLAAPVGATLGTSTSAMGTINNDD1069 pKa = 3.1DD1070 pKa = 4.1TLPTINVAPVYY1081 pKa = 10.55AITEE1085 pKa = 4.44GNSGSTTVNVTVNRR1099 pKa = 11.84TGDD1102 pKa = 3.34LSTASSANWSVAGSGTTPAIGADD1125 pKa = 3.59FTGGVLPTGTVNFAAGIATASIAINVAGDD1154 pKa = 3.59TVFEE1158 pKa = 4.22QNEE1161 pKa = 4.22NFTVSLNTPVNATLGASVNTTGTINNDD1188 pKa = 2.92DD1189 pKa = 4.27TLPVLNVAPVLAITEE1204 pKa = 4.43GNSGSTAVNITVNRR1218 pKa = 11.84TGDD1221 pKa = 3.44LSTASSTNWSVAGSGTNPVIGADD1244 pKa = 3.57FTGGVLPTGTVNFAAGVATATIAINVAGDD1273 pKa = 3.43TTFEE1277 pKa = 4.44PNEE1280 pKa = 4.32DD1281 pKa = 3.49FTVSLNTPVNATLDD1295 pKa = 3.54ASVNTTGTINNDD1307 pKa = 2.92DD1308 pKa = 4.27TLPVLNVAPVLAITEE1323 pKa = 4.43GNSGSTAVNVTVNRR1337 pKa = 11.84TGDD1340 pKa = 3.34LSTASSANWSVAGSGTNPVIGADD1363 pKa = 3.62FMGGVLPMGTVNFAAGLATATIAINVAGDD1392 pKa = 3.43TTFEE1396 pKa = 4.44PNEE1399 pKa = 4.32DD1400 pKa = 3.49FTVSLNTPVNATLDD1414 pKa = 3.54ASVNTTGTINNDD1426 pKa = 2.92DD1427 pKa = 4.27TLPVLNVAPVLAITEE1442 pKa = 4.43GNSGSTAVNVTVNRR1456 pKa = 11.84TGDD1459 pKa = 3.34LSTASSANWSVAGSGTNPAIGADD1482 pKa = 3.7FTGGVLPTGMVNFAAGIATASIAINVAGDD1511 pKa = 3.59TVFEE1515 pKa = 4.22QNEE1518 pKa = 4.22NFTVSLNTPVNATLGSSVNTTGTINNDD1545 pKa = 2.92DD1546 pKa = 4.27TLPVLNVAPVLAITEE1561 pKa = 4.43GNSGSTAVNITVNRR1575 pKa = 11.84TGDD1578 pKa = 3.36LSTASSANWSVAGSGTNPVIGADD1601 pKa = 3.57FTGGVLPTGTVNFAAGVATATIAINVAGDD1630 pKa = 3.43TLFEE1634 pKa = 4.32QNEE1637 pKa = 4.25NFTVSLNTPVNATLGTSTSAMGTINNDD1664 pKa = 2.95DD1665 pKa = 4.23SIAIVGTSNNDD1676 pKa = 3.34RR1677 pKa = 11.84LKK1679 pKa = 10.71GTIKK1683 pKa = 10.4SDD1685 pKa = 3.58QLDD1688 pKa = 3.75GLAGDD1693 pKa = 5.21DD1694 pKa = 3.9YY1695 pKa = 11.65FFGLGGNDD1703 pKa = 3.66TLNGGAGNDD1712 pKa = 3.59TMRR1715 pKa = 11.84GGTGLDD1721 pKa = 3.49SMIGGDD1727 pKa = 4.77GNDD1730 pKa = 3.19LYY1732 pKa = 11.37YY1733 pKa = 10.65VEE1735 pKa = 5.42EE1736 pKa = 5.26IGDD1739 pKa = 3.88IVVEE1743 pKa = 4.27TATGGIDD1750 pKa = 3.45TVNSSLNNYY1759 pKa = 7.0MLNAAVEE1766 pKa = 4.26NGRR1769 pKa = 11.84ISIPGTASITGNDD1782 pKa = 3.67LDD1784 pKa = 4.91NILFAGVGNNTIDD1797 pKa = 3.7GGTGTDD1803 pKa = 3.15TVVYY1807 pKa = 9.77SALKK1811 pKa = 8.87TSVGSTLGVKK1821 pKa = 9.92VNLMNTAIQITGSSGSDD1838 pKa = 2.99LLLSIEE1844 pKa = 4.53NLTGSNKK1851 pKa = 9.84NDD1853 pKa = 3.56TLGGNSAANVLNGGLGNDD1871 pKa = 3.76ILTGGAGKK1879 pKa = 10.23DD1880 pKa = 3.11VFVFDD1885 pKa = 3.5TALNRR1890 pKa = 11.84NVDD1893 pKa = 3.74QIKK1896 pKa = 10.67DD1897 pKa = 3.58FSVVDD1902 pKa = 3.58DD1903 pKa = 4.92TIQLEE1908 pKa = 4.17NSVFKK1913 pKa = 10.78QLSSTGSLKK1922 pKa = 10.76ASYY1925 pKa = 9.44FVKK1928 pKa = 9.85NTTGKK1933 pKa = 10.79ANDD1936 pKa = 3.59TDD1938 pKa = 4.54DD1939 pKa = 5.21FIIYY1943 pKa = 8.16DD1944 pKa = 3.98TDD1946 pKa = 3.63SGALFYY1952 pKa = 11.19DD1953 pKa = 3.96ADD1955 pKa = 4.01GSGAGAAVQFAIVSIGVVLTAADD1978 pKa = 4.96FIVSS1982 pKa = 3.71
MM1 pKa = 6.83FASRR5 pKa = 11.84AHH7 pKa = 6.18AVLITDD13 pKa = 4.73AQLQDD18 pKa = 4.44LDD20 pKa = 3.85QLLAGLDD27 pKa = 3.5ADD29 pKa = 3.99VEE31 pKa = 4.38PWLVASNQDD40 pKa = 3.28AMPLIFKK47 pKa = 10.64ALAHH51 pKa = 6.95PDD53 pKa = 3.39LTQLHH58 pKa = 6.79LLAHH62 pKa = 6.69GAPGEE67 pKa = 3.96IRR69 pKa = 11.84LGEE72 pKa = 4.17RR73 pKa = 11.84ALTAADD79 pKa = 3.6FRR81 pKa = 11.84QYY83 pKa = 11.5LNGAAMRR90 pKa = 11.84DD91 pKa = 3.42LSINFWSCHH100 pKa = 4.94TGAGEE105 pKa = 3.87MGTEE109 pKa = 4.24FTHH112 pKa = 6.96AVAAATGARR121 pKa = 11.84VLAAEE126 pKa = 4.31GLIGAANKK134 pKa = 9.52QGSWEE139 pKa = 4.23LNGITAPFSEE149 pKa = 4.82TARR152 pKa = 11.84AGYY155 pKa = 11.06ANVLAVRR162 pKa = 11.84DD163 pKa = 3.86RR164 pKa = 11.84FEE166 pKa = 6.41DD167 pKa = 3.39NDD169 pKa = 3.98TAATATNLGTITSTRR184 pKa = 11.84TEE186 pKa = 3.99TGLSIEE192 pKa = 4.66TNDD195 pKa = 3.85PDD197 pKa = 3.39WFQFTLGSTGTSASNVAIDD216 pKa = 3.96FSNALGDD223 pKa = 4.36LDD225 pKa = 4.21IFLYY229 pKa = 10.76DD230 pKa = 3.32AAGNALQGTYY240 pKa = 10.42GSNDD244 pKa = 3.22HH245 pKa = 6.04EE246 pKa = 4.61QISLSGLAAGSYY258 pKa = 9.66LIKK261 pKa = 10.85VRR263 pKa = 11.84DD264 pKa = 3.6WTSLDD269 pKa = 3.22GGEE272 pKa = 4.28NPNYY276 pKa = 9.45TLTLIGSGEE285 pKa = 4.03VSPTVSYY292 pKa = 7.64PTLDD296 pKa = 3.51YY297 pKa = 10.98MPDD300 pKa = 3.37LTSSQMSGARR310 pKa = 11.84SLTFAPYY317 pKa = 10.25TSTSWDD323 pKa = 3.59SKK325 pKa = 9.78AGAYY329 pKa = 10.06GSEE332 pKa = 4.46SGASSHH338 pKa = 7.05SIYY341 pKa = 10.43TITATAGQVYY351 pKa = 10.44DD352 pKa = 5.02LYY354 pKa = 11.03SVSYY358 pKa = 9.78FDD360 pKa = 4.79PYY362 pKa = 11.5SLTVYY367 pKa = 10.59DD368 pKa = 4.15SLGNAIVTNAEE379 pKa = 3.69SDD381 pKa = 3.82DD382 pKa = 3.94PADD385 pKa = 3.8YY386 pKa = 11.43NLGDD390 pKa = 3.77AYY392 pKa = 11.47YY393 pKa = 10.88GVDD396 pKa = 3.93WLNDD400 pKa = 2.87WTAPYY405 pKa = 10.47SGTYY409 pKa = 9.58YY410 pKa = 10.93VDD412 pKa = 4.35ASWHH416 pKa = 5.08QGSYY420 pKa = 7.75FTFYY424 pKa = 11.62DD425 pKa = 3.9LMIYY429 pKa = 9.93KK430 pKa = 10.29NKK432 pKa = 9.06STTVLPTLSLSSLSPSSVSEE452 pKa = 4.22GNSGSTSMMVNVTRR466 pKa = 11.84TGDD469 pKa = 3.45LSSTSSASWSVVGAAATASDD489 pKa = 4.27FLSGVLPTGTVNFAAGQASATATINIAGDD518 pKa = 3.46TTIEE522 pKa = 3.84SDD524 pKa = 3.76EE525 pKa = 4.39NFTVTLSNPVSATLDD540 pKa = 3.16ISSATGTIVNDD551 pKa = 3.98DD552 pKa = 3.61VSPPVPYY559 pKa = 8.19PTLDD563 pKa = 3.75YY564 pKa = 11.1IPDD567 pKa = 3.97LKK569 pKa = 10.8SSQMYY574 pKa = 9.81EE575 pKa = 3.75ADD577 pKa = 4.08LLAFLPVTSSVYY589 pKa = 10.47YY590 pKa = 10.92DD591 pKa = 3.43MEE593 pKa = 6.61AYY595 pKa = 10.15DD596 pKa = 4.9YY597 pKa = 11.29GLTSNASLHH606 pKa = 6.44SLYY609 pKa = 10.62QFNATTGVLYY619 pKa = 10.32DD620 pKa = 4.35IYY622 pKa = 10.65SISYY626 pKa = 10.41LDD628 pKa = 3.63PTEE631 pKa = 3.93LTIYY635 pKa = 10.41DD636 pKa = 3.6QFGNAIAINNEE647 pKa = 3.52SDD649 pKa = 4.64DD650 pKa = 4.42PDD652 pKa = 3.7NSTLSWVDD660 pKa = 3.36DD661 pKa = 3.99GGSYY665 pKa = 10.79SFDD668 pKa = 5.43AIWEE672 pKa = 3.95WSAPYY677 pKa = 10.47SGTFYY682 pKa = 11.24VDD684 pKa = 3.34SGWNQGSFFTYY695 pKa = 9.14WDD697 pKa = 3.25VSVYY701 pKa = 10.25RR702 pKa = 11.84NKK704 pKa = 9.91STTPSLPTLSLSSLTPTSISEE725 pKa = 4.32GNSGYY730 pKa = 9.0TPITFTVNRR739 pKa = 11.84DD740 pKa = 2.9GDD742 pKa = 3.73LSGTSRR748 pKa = 11.84VFFAVTGTGTNPVNAADD765 pKa = 4.08FRR767 pKa = 11.84DD768 pKa = 4.06GVLPSDD774 pKa = 4.47WIDD777 pKa = 3.57FDD779 pKa = 4.13SGAVSATITSYY790 pKa = 11.3VVAGDD795 pKa = 3.59TTPEE799 pKa = 3.95PNEE802 pKa = 4.03NFTVTLSSPTGATLGTSTSVIGTITNDD829 pKa = 3.6DD830 pKa = 4.35SLPLPTISLSPLSPSIITEE849 pKa = 3.91GHH851 pKa = 5.52SGYY854 pKa = 8.97TPVNIVVTRR863 pKa = 11.84GGDD866 pKa = 3.41LSSTSSATWTVVGTGVAPVTADD888 pKa = 3.45DD889 pKa = 4.54FGGFLPSGIISLAAGVATATVSINILGDD917 pKa = 3.39TTVEE921 pKa = 4.18SNEE924 pKa = 4.54DD925 pKa = 3.49FTVMLSNPSGATLIPNTVTGTITNDD950 pKa = 3.32DD951 pKa = 4.28VILPTLSLASLTPSIIMEE969 pKa = 4.48GNSGFTPMTVTITRR983 pKa = 11.84SGDD986 pKa = 3.4LSGTSAANWSVIGAGATADD1005 pKa = 3.81DD1006 pKa = 4.95FIGGVLPSGTISFPAWQATTTATINIFGDD1035 pKa = 3.62TLVEE1039 pKa = 4.38SNEE1042 pKa = 3.95NFSVVLAAPVGATLGTSTSAMGTINNDD1069 pKa = 3.1DD1070 pKa = 4.1TLPTINVAPVYY1081 pKa = 10.55AITEE1085 pKa = 4.44GNSGSTTVNVTVNRR1099 pKa = 11.84TGDD1102 pKa = 3.34LSTASSANWSVAGSGTTPAIGADD1125 pKa = 3.59FTGGVLPTGTVNFAAGIATASIAINVAGDD1154 pKa = 3.59TVFEE1158 pKa = 4.22QNEE1161 pKa = 4.22NFTVSLNTPVNATLGASVNTTGTINNDD1188 pKa = 2.92DD1189 pKa = 4.27TLPVLNVAPVLAITEE1204 pKa = 4.43GNSGSTAVNITVNRR1218 pKa = 11.84TGDD1221 pKa = 3.44LSTASSTNWSVAGSGTNPVIGADD1244 pKa = 3.57FTGGVLPTGTVNFAAGVATATIAINVAGDD1273 pKa = 3.43TTFEE1277 pKa = 4.44PNEE1280 pKa = 4.32DD1281 pKa = 3.49FTVSLNTPVNATLDD1295 pKa = 3.54ASVNTTGTINNDD1307 pKa = 2.92DD1308 pKa = 4.27TLPVLNVAPVLAITEE1323 pKa = 4.43GNSGSTAVNVTVNRR1337 pKa = 11.84TGDD1340 pKa = 3.34LSTASSANWSVAGSGTNPVIGADD1363 pKa = 3.62FMGGVLPMGTVNFAAGLATATIAINVAGDD1392 pKa = 3.43TTFEE1396 pKa = 4.44PNEE1399 pKa = 4.32DD1400 pKa = 3.49FTVSLNTPVNATLDD1414 pKa = 3.54ASVNTTGTINNDD1426 pKa = 2.92DD1427 pKa = 4.27TLPVLNVAPVLAITEE1442 pKa = 4.43GNSGSTAVNVTVNRR1456 pKa = 11.84TGDD1459 pKa = 3.34LSTASSANWSVAGSGTNPAIGADD1482 pKa = 3.7FTGGVLPTGMVNFAAGIATASIAINVAGDD1511 pKa = 3.59TVFEE1515 pKa = 4.22QNEE1518 pKa = 4.22NFTVSLNTPVNATLGSSVNTTGTINNDD1545 pKa = 2.92DD1546 pKa = 4.27TLPVLNVAPVLAITEE1561 pKa = 4.43GNSGSTAVNITVNRR1575 pKa = 11.84TGDD1578 pKa = 3.36LSTASSANWSVAGSGTNPVIGADD1601 pKa = 3.57FTGGVLPTGTVNFAAGVATATIAINVAGDD1630 pKa = 3.43TLFEE1634 pKa = 4.32QNEE1637 pKa = 4.25NFTVSLNTPVNATLGTSTSAMGTINNDD1664 pKa = 2.95DD1665 pKa = 4.23SIAIVGTSNNDD1676 pKa = 3.34RR1677 pKa = 11.84LKK1679 pKa = 10.71GTIKK1683 pKa = 10.4SDD1685 pKa = 3.58QLDD1688 pKa = 3.75GLAGDD1693 pKa = 5.21DD1694 pKa = 3.9YY1695 pKa = 11.65FFGLGGNDD1703 pKa = 3.66TLNGGAGNDD1712 pKa = 3.59TMRR1715 pKa = 11.84GGTGLDD1721 pKa = 3.49SMIGGDD1727 pKa = 4.77GNDD1730 pKa = 3.19LYY1732 pKa = 11.37YY1733 pKa = 10.65VEE1735 pKa = 5.42EE1736 pKa = 5.26IGDD1739 pKa = 3.88IVVEE1743 pKa = 4.27TATGGIDD1750 pKa = 3.45TVNSSLNNYY1759 pKa = 7.0MLNAAVEE1766 pKa = 4.26NGRR1769 pKa = 11.84ISIPGTASITGNDD1782 pKa = 3.67LDD1784 pKa = 4.91NILFAGVGNNTIDD1797 pKa = 3.7GGTGTDD1803 pKa = 3.15TVVYY1807 pKa = 9.77SALKK1811 pKa = 8.87TSVGSTLGVKK1821 pKa = 9.92VNLMNTAIQITGSSGSDD1838 pKa = 2.99LLLSIEE1844 pKa = 4.53NLTGSNKK1851 pKa = 9.84NDD1853 pKa = 3.56TLGGNSAANVLNGGLGNDD1871 pKa = 3.76ILTGGAGKK1879 pKa = 10.23DD1880 pKa = 3.11VFVFDD1885 pKa = 3.5TALNRR1890 pKa = 11.84NVDD1893 pKa = 3.74QIKK1896 pKa = 10.67DD1897 pKa = 3.58FSVVDD1902 pKa = 3.58DD1903 pKa = 4.92TIQLEE1908 pKa = 4.17NSVFKK1913 pKa = 10.78QLSSTGSLKK1922 pKa = 10.76ASYY1925 pKa = 9.44FVKK1928 pKa = 9.85NTTGKK1933 pKa = 10.79ANDD1936 pKa = 3.59TDD1938 pKa = 4.54DD1939 pKa = 5.21FIIYY1943 pKa = 8.16DD1944 pKa = 3.98TDD1946 pKa = 3.63SGALFYY1952 pKa = 11.19DD1953 pKa = 3.96ADD1955 pKa = 4.01GSGAGAAVQFAIVSIGVVLTAADD1978 pKa = 4.96FIVSS1982 pKa = 3.71
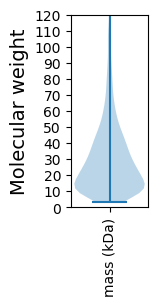
Molecular weight: 203.03 kDa
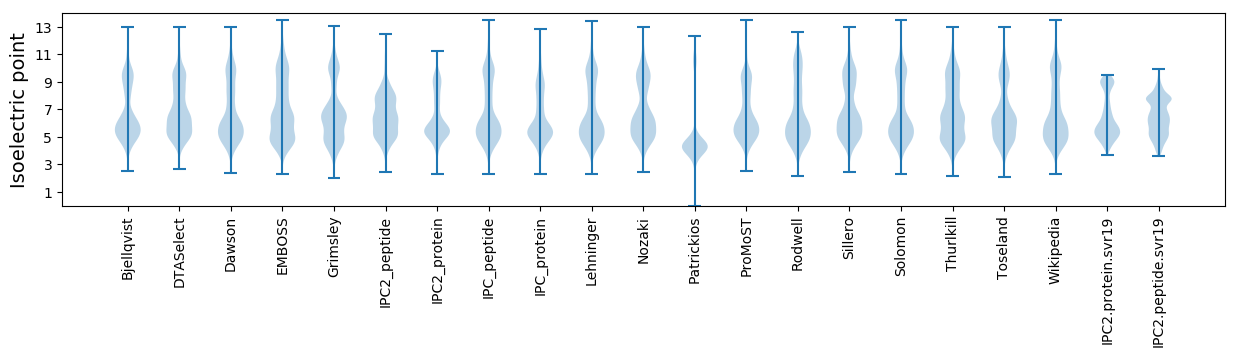
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S7XQK1|A0A2S7XQK1_9GAMM 50S ribosomal protein L14 OS=Chromatium okenii OX=61644 GN=rplN PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84SATRR25 pKa = 11.84NGLKK29 pKa = 10.3VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSPVV45 pKa = 2.69
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84SATRR25 pKa = 11.84NGLKK29 pKa = 10.3VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSPVV45 pKa = 2.69
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
814741 |
27 |
2927 |
289.8 |
31.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.737 ± 0.068 | 1.148 ± 0.027 |
5.518 ± 0.039 | 5.656 ± 0.05 |
3.767 ± 0.036 | 6.88 ± 0.06 |
2.311 ± 0.028 | 5.922 ± 0.04 |
3.571 ± 0.045 | 10.952 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.22 ± 0.027 | 3.856 ± 0.056 |
4.67 ± 0.051 | 4.471 ± 0.044 |
6.261 ± 0.065 | 5.505 ± 0.054 |
6.073 ± 0.087 | 6.686 ± 0.044 |
1.316 ± 0.022 | 2.479 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
