
West African Asystasia virus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 7.91
Get precalculated fractions of proteins
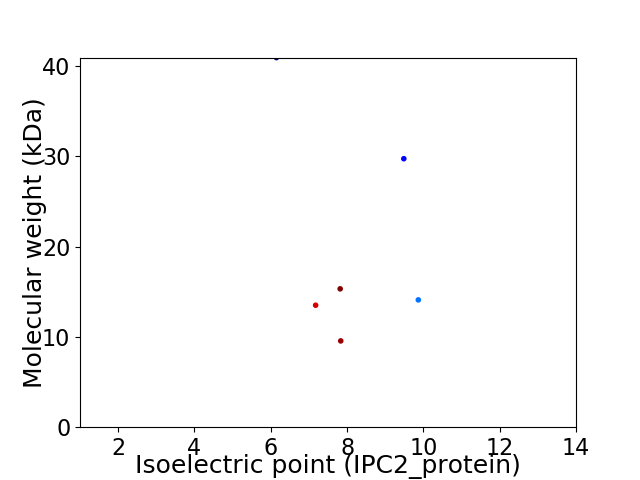
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G9CM36|G9CM36_9GEMI AC4 protein OS=West African Asystasia virus 2 OX=1046574 GN=AC4 PE=3 SV=1
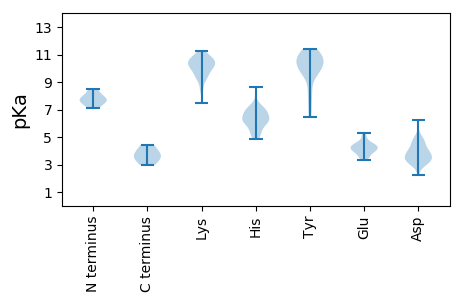
MM1 pKa = 7.65APPKK5 pKa = 10.4RR6 pKa = 11.84FLINCKK12 pKa = 10.28NYY14 pKa = 9.78FLTYY18 pKa = 8.41PQCPLTKK25 pKa = 10.33EE26 pKa = 3.99EE27 pKa = 4.77ALSQIINLSTPTNKK41 pKa = 10.01LFIKK45 pKa = 10.0ICRR48 pKa = 11.84EE49 pKa = 3.52LHH51 pKa = 6.26EE52 pKa = 4.93NGEE55 pKa = 4.41PHH57 pKa = 7.12LHH59 pKa = 6.03MLVQFEE65 pKa = 5.29GKK67 pKa = 9.97FKK69 pKa = 10.6CQNKK73 pKa = 9.52RR74 pKa = 11.84FFDD77 pKa = 3.9LVSPTRR83 pKa = 11.84SAHH86 pKa = 4.86FHH88 pKa = 6.55PNIQGAKK95 pKa = 9.25SSSDD99 pKa = 3.06VKK101 pKa = 11.23SYY103 pKa = 10.83IEE105 pKa = 4.16KK106 pKa = 10.94DD107 pKa = 3.04GDD109 pKa = 3.7TLEE112 pKa = 4.34WGTFQIDD119 pKa = 3.03GRR121 pKa = 11.84SARR124 pKa = 11.84GGCQNANDD132 pKa = 4.04ACAEE136 pKa = 3.94ALNAGSADD144 pKa = 3.29AALAIIRR151 pKa = 11.84EE152 pKa = 4.23KK153 pKa = 10.87LPKK156 pKa = 10.46DD157 pKa = 4.77FIFQYY162 pKa = 11.18HH163 pKa = 5.38NLKK166 pKa = 10.92SNLDD170 pKa = 4.37RR171 pKa = 11.84IFMSPPEE178 pKa = 4.46AYY180 pKa = 9.95VSPFLSSSFDD190 pKa = 3.44QVPEE194 pKa = 3.87EE195 pKa = 4.25LEE197 pKa = 4.08VWVSEE202 pKa = 4.06NVMGPAARR210 pKa = 11.84PWRR213 pKa = 11.84PNSIVIEE220 pKa = 4.34GDD222 pKa = 3.08SRR224 pKa = 11.84TGKK227 pKa = 8.52TMWARR232 pKa = 11.84SLGPHH237 pKa = 6.71NYY239 pKa = 10.18LCGHH243 pKa = 7.38LDD245 pKa = 4.18LSPKK249 pKa = 10.15VYY251 pKa = 11.07SNDD254 pKa = 2.25AWYY257 pKa = 10.92NVIDD261 pKa = 5.2DD262 pKa = 4.41VDD264 pKa = 3.61PHH266 pKa = 5.85YY267 pKa = 11.0LKK269 pKa = 10.7HH270 pKa = 6.2FKK272 pKa = 10.7EE273 pKa = 4.4FMGAQRR279 pKa = 11.84NWNSNVKK286 pKa = 9.8YY287 pKa = 10.24GKK289 pKa = 9.2PVQIKK294 pKa = 10.41GGIPTIFLCNPGPTSSYY311 pKa = 10.9KK312 pKa = 10.53EE313 pKa = 3.85YY314 pKa = 11.17LEE316 pKa = 4.21EE317 pKa = 4.38EE318 pKa = 4.59KK319 pKa = 11.1NSSLKK324 pKa = 9.25TWAIKK329 pKa = 9.98NASFITLVGPLYY341 pKa = 10.93SGANQSQPQTRR352 pKa = 11.84EE353 pKa = 3.86TQTNPQEE360 pKa = 3.98EE361 pKa = 4.57DD362 pKa = 2.96
MM1 pKa = 7.65APPKK5 pKa = 10.4RR6 pKa = 11.84FLINCKK12 pKa = 10.28NYY14 pKa = 9.78FLTYY18 pKa = 8.41PQCPLTKK25 pKa = 10.33EE26 pKa = 3.99EE27 pKa = 4.77ALSQIINLSTPTNKK41 pKa = 10.01LFIKK45 pKa = 10.0ICRR48 pKa = 11.84EE49 pKa = 3.52LHH51 pKa = 6.26EE52 pKa = 4.93NGEE55 pKa = 4.41PHH57 pKa = 7.12LHH59 pKa = 6.03MLVQFEE65 pKa = 5.29GKK67 pKa = 9.97FKK69 pKa = 10.6CQNKK73 pKa = 9.52RR74 pKa = 11.84FFDD77 pKa = 3.9LVSPTRR83 pKa = 11.84SAHH86 pKa = 4.86FHH88 pKa = 6.55PNIQGAKK95 pKa = 9.25SSSDD99 pKa = 3.06VKK101 pKa = 11.23SYY103 pKa = 10.83IEE105 pKa = 4.16KK106 pKa = 10.94DD107 pKa = 3.04GDD109 pKa = 3.7TLEE112 pKa = 4.34WGTFQIDD119 pKa = 3.03GRR121 pKa = 11.84SARR124 pKa = 11.84GGCQNANDD132 pKa = 4.04ACAEE136 pKa = 3.94ALNAGSADD144 pKa = 3.29AALAIIRR151 pKa = 11.84EE152 pKa = 4.23KK153 pKa = 10.87LPKK156 pKa = 10.46DD157 pKa = 4.77FIFQYY162 pKa = 11.18HH163 pKa = 5.38NLKK166 pKa = 10.92SNLDD170 pKa = 4.37RR171 pKa = 11.84IFMSPPEE178 pKa = 4.46AYY180 pKa = 9.95VSPFLSSSFDD190 pKa = 3.44QVPEE194 pKa = 3.87EE195 pKa = 4.25LEE197 pKa = 4.08VWVSEE202 pKa = 4.06NVMGPAARR210 pKa = 11.84PWRR213 pKa = 11.84PNSIVIEE220 pKa = 4.34GDD222 pKa = 3.08SRR224 pKa = 11.84TGKK227 pKa = 8.52TMWARR232 pKa = 11.84SLGPHH237 pKa = 6.71NYY239 pKa = 10.18LCGHH243 pKa = 7.38LDD245 pKa = 4.18LSPKK249 pKa = 10.15VYY251 pKa = 11.07SNDD254 pKa = 2.25AWYY257 pKa = 10.92NVIDD261 pKa = 5.2DD262 pKa = 4.41VDD264 pKa = 3.61PHH266 pKa = 5.85YY267 pKa = 11.0LKK269 pKa = 10.7HH270 pKa = 6.2FKK272 pKa = 10.7EE273 pKa = 4.4FMGAQRR279 pKa = 11.84NWNSNVKK286 pKa = 9.8YY287 pKa = 10.24GKK289 pKa = 9.2PVQIKK294 pKa = 10.41GGIPTIFLCNPGPTSSYY311 pKa = 10.9KK312 pKa = 10.53EE313 pKa = 3.85YY314 pKa = 11.17LEE316 pKa = 4.21EE317 pKa = 4.38EE318 pKa = 4.59KK319 pKa = 11.1NSSLKK324 pKa = 9.25TWAIKK329 pKa = 9.98NASFITLVGPLYY341 pKa = 10.93SGANQSQPQTRR352 pKa = 11.84EE353 pKa = 3.86TQTNPQEE360 pKa = 3.98EE361 pKa = 4.57DD362 pKa = 2.96
Molecular weight: 40.93 kDa
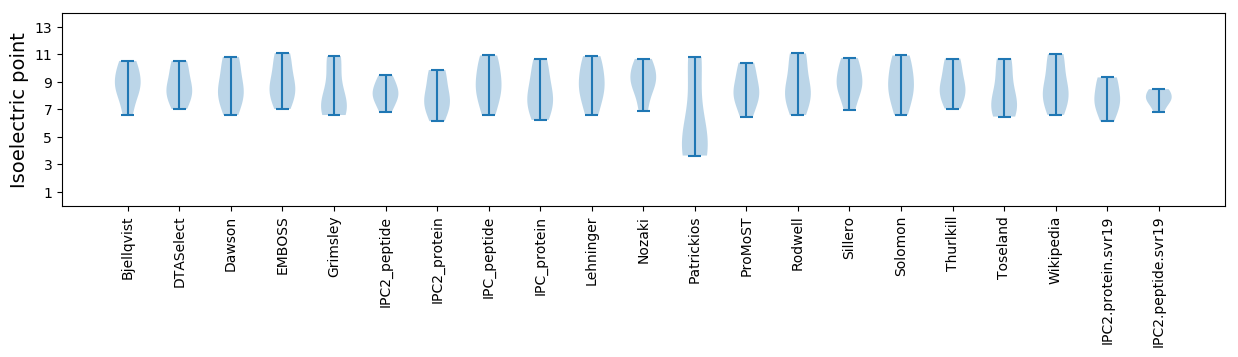
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9CM34|G9CM34_9GEMI Transcriptional activator protein OS=West African Asystasia virus 2 OX=1046574 GN=AC2 PE=3 SV=1
MM1 pKa = 8.51DD2 pKa = 4.64SRR4 pKa = 11.84TEE6 pKa = 3.97EE7 pKa = 4.41NITAAQAKK15 pKa = 8.49NGVYY19 pKa = 9.4IWEE22 pKa = 4.4ISNPLYY28 pKa = 10.66FKK30 pKa = 10.6ILNHH34 pKa = 6.29DD35 pKa = 4.05VRR37 pKa = 11.84PMSRR41 pKa = 11.84PWDD44 pKa = 3.64IIKK47 pKa = 10.04IQVRR51 pKa = 11.84FNHH54 pKa = 6.14NLRR57 pKa = 11.84KK58 pKa = 9.81ALGIHH63 pKa = 6.15RR64 pKa = 11.84CLLTFQAWTSLRR76 pKa = 11.84PQTSHH81 pKa = 7.05FLRR84 pKa = 11.84VFSKK88 pKa = 10.83HH89 pKa = 4.94VIRR92 pKa = 11.84YY93 pKa = 8.0LNSLGVLSINNLIRR107 pKa = 11.84AIKK110 pKa = 9.31FVLYY114 pKa = 10.53NVLHH118 pKa = 5.62GTT120 pKa = 3.73
MM1 pKa = 8.51DD2 pKa = 4.64SRR4 pKa = 11.84TEE6 pKa = 3.97EE7 pKa = 4.41NITAAQAKK15 pKa = 8.49NGVYY19 pKa = 9.4IWEE22 pKa = 4.4ISNPLYY28 pKa = 10.66FKK30 pKa = 10.6ILNHH34 pKa = 6.29DD35 pKa = 4.05VRR37 pKa = 11.84PMSRR41 pKa = 11.84PWDD44 pKa = 3.64IIKK47 pKa = 10.04IQVRR51 pKa = 11.84FNHH54 pKa = 6.14NLRR57 pKa = 11.84KK58 pKa = 9.81ALGIHH63 pKa = 6.15RR64 pKa = 11.84CLLTFQAWTSLRR76 pKa = 11.84PQTSHH81 pKa = 7.05FLRR84 pKa = 11.84VFSKK88 pKa = 10.83HH89 pKa = 4.94VIRR92 pKa = 11.84YY93 pKa = 8.0LNSLGVLSINNLIRR107 pKa = 11.84AIKK110 pKa = 9.31FVLYY114 pKa = 10.53NVLHH118 pKa = 5.62GTT120 pKa = 3.73
Molecular weight: 14.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1075 |
85 |
362 |
179.2 |
20.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.395 ± 0.782 | 2.512 ± 0.499 |
4.651 ± 0.399 | 4.744 ± 0.864 |
4.279 ± 0.395 | 5.209 ± 0.575 |
4.0 ± 0.653 | 5.488 ± 0.845 |
6.047 ± 0.615 | 7.814 ± 0.734 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.884 ± 0.68 | 6.233 ± 0.503 |
6.419 ± 0.641 | 4.372 ± 0.555 |
6.791 ± 1.165 | 7.814 ± 1.132 |
5.116 ± 0.268 | 5.209 ± 0.79 |
1.395 ± 0.282 | 3.628 ± 0.555 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
