
Nematocida displodere
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Microsporidia; Microsporidia incertae sedis; Nematocida
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
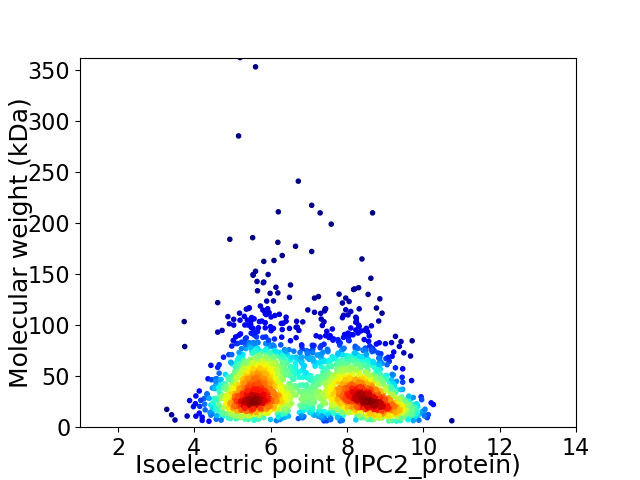
Virtual 2D-PAGE plot for 2241 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177EAE3|A0A177EAE3_9MICR Uncharacterized protein OS=Nematocida displodere OX=1805483 GN=NEDG_01024 PE=4 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84RR3 pKa = 11.84QAQHH7 pKa = 5.66RR8 pKa = 11.84CKK10 pKa = 10.59AILLLASLYY19 pKa = 9.31VLCGCIEE26 pKa = 4.18IDD28 pKa = 3.47PVVISTLEE36 pKa = 4.08DD37 pKa = 3.33FAKK40 pKa = 10.68DD41 pKa = 3.64LNEE44 pKa = 5.32KK45 pKa = 8.73PTIDD49 pKa = 3.09QNMFDD54 pKa = 4.47EE55 pKa = 5.26EE56 pKa = 4.27IKK58 pKa = 11.09ALTEE62 pKa = 3.92AKK64 pKa = 10.36LATEE68 pKa = 4.27EE69 pKa = 5.21KK70 pKa = 10.02IDD72 pKa = 3.87LLKK75 pKa = 10.77QRR77 pKa = 11.84GVQSGLATEE86 pKa = 4.6EE87 pKa = 4.48SNVPEE92 pKa = 4.19EE93 pKa = 3.92QSPAEE98 pKa = 4.03EE99 pKa = 3.76QLQAEE104 pKa = 4.57EE105 pKa = 4.06LAIGNDD111 pKa = 3.49EE112 pKa = 4.02QSEE115 pKa = 4.43VEE117 pKa = 4.17PEE119 pKa = 4.09VEE121 pKa = 4.05EE122 pKa = 4.36EE123 pKa = 3.97PTVFDD128 pKa = 3.64EE129 pKa = 4.01QSEE132 pKa = 4.55YY133 pKa = 11.06EE134 pKa = 4.09EE135 pKa = 4.41EE136 pKa = 4.07EE137 pKa = 4.19QYY139 pKa = 10.84EE140 pKa = 4.33AEE142 pKa = 4.52PATEE146 pKa = 3.74PTVFDD151 pKa = 3.66EE152 pKa = 4.29QLEE155 pKa = 4.35EE156 pKa = 4.22EE157 pKa = 4.87YY158 pKa = 10.69EE159 pKa = 4.32AEE161 pKa = 4.26PEE163 pKa = 4.06AADD166 pKa = 3.49EE167 pKa = 4.64AEE169 pKa = 4.11YY170 pKa = 10.9DD171 pKa = 3.88GQPEE175 pKa = 4.22EE176 pKa = 4.26QYY178 pKa = 10.53EE179 pKa = 4.12AQPEE183 pKa = 4.22PEE185 pKa = 4.25AEE187 pKa = 3.94EE188 pKa = 4.46EE189 pKa = 4.38YY190 pKa = 10.84EE191 pKa = 4.29PEE193 pKa = 4.48AEE195 pKa = 4.16PEE197 pKa = 3.97AEE199 pKa = 4.31EE200 pKa = 5.22EE201 pKa = 4.12YY202 pKa = 10.86DD203 pKa = 3.77GQPEE207 pKa = 4.09EE208 pKa = 4.21QYY210 pKa = 10.66EE211 pKa = 4.44AEE213 pKa = 4.46PEE215 pKa = 3.92AADD218 pKa = 4.44GIEE221 pKa = 3.79YY222 pKa = 10.43DD223 pKa = 3.92GQSEE227 pKa = 4.37EE228 pKa = 4.18QYY230 pKa = 10.2EE231 pKa = 4.24AQPEE235 pKa = 4.22AEE237 pKa = 5.05AEE239 pKa = 4.13AEE241 pKa = 4.21EE242 pKa = 4.15QSEE245 pKa = 4.39PEE247 pKa = 4.2EE248 pKa = 4.13QPEE251 pKa = 4.08AEE253 pKa = 4.17PEE255 pKa = 4.22VFDD258 pKa = 4.25EE259 pKa = 4.21QLEE262 pKa = 4.35AEE264 pKa = 4.66PEE266 pKa = 4.12PEE268 pKa = 4.17PEE270 pKa = 5.17AEE272 pKa = 4.37AEE274 pKa = 3.88YY275 pKa = 10.85DD276 pKa = 3.94GQSEE280 pKa = 4.61EE281 pKa = 3.98QDD283 pKa = 3.15EE284 pKa = 4.57AQPEE288 pKa = 4.44TQLEE292 pKa = 4.25AEE294 pKa = 4.43PEE296 pKa = 3.99AEE298 pKa = 4.22EE299 pKa = 5.15EE300 pKa = 4.01YY301 pKa = 10.86DD302 pKa = 3.79GQSEE306 pKa = 4.18EE307 pKa = 4.22QYY309 pKa = 10.84EE310 pKa = 4.38PEE312 pKa = 4.31AQPEE316 pKa = 4.17AEE318 pKa = 4.31PEE320 pKa = 3.96AEE322 pKa = 4.21AEE324 pKa = 4.48AEE326 pKa = 4.05AEE328 pKa = 3.84YY329 pKa = 10.87DD330 pKa = 3.83GQSEE334 pKa = 4.44EE335 pKa = 4.18QYY337 pKa = 9.98EE338 pKa = 4.34AQPEE342 pKa = 4.22TEE344 pKa = 3.76PAVFDD349 pKa = 3.96EE350 pKa = 4.26QLEE353 pKa = 4.34AEE355 pKa = 4.59PEE357 pKa = 4.11AEE359 pKa = 4.16PEE361 pKa = 4.08AEE363 pKa = 4.58PEE365 pKa = 4.49AEE367 pKa = 4.24TQLEE371 pKa = 4.24AEE373 pKa = 4.62PEE375 pKa = 4.06AEE377 pKa = 4.2AEE379 pKa = 4.48AEE381 pKa = 4.05AEE383 pKa = 3.84YY384 pKa = 10.87DD385 pKa = 3.83GQSEE389 pKa = 4.3EE390 pKa = 4.17QYY392 pKa = 10.24EE393 pKa = 4.25AEE395 pKa = 4.78PEE397 pKa = 4.14PEE399 pKa = 4.22TQLEE403 pKa = 4.34AEE405 pKa = 4.62PEE407 pKa = 4.09AEE409 pKa = 4.27PEE411 pKa = 4.2AEE413 pKa = 4.28ADD415 pKa = 3.37YY416 pKa = 11.23DD417 pKa = 3.97GQSEE421 pKa = 4.61YY422 pKa = 10.56EE423 pKa = 3.83AEE425 pKa = 4.0YY426 pKa = 10.9DD427 pKa = 4.57GEE429 pKa = 4.79SEE431 pKa = 4.26TQLEE435 pKa = 4.24EE436 pKa = 4.09EE437 pKa = 4.4EE438 pKa = 4.36QYY440 pKa = 10.85EE441 pKa = 4.37AEE443 pKa = 4.53PEE445 pKa = 3.96PTVYY449 pKa = 10.62DD450 pKa = 3.82EE451 pKa = 4.23QLEE454 pKa = 4.37AEE456 pKa = 4.58PEE458 pKa = 4.07AEE460 pKa = 3.97PEE462 pKa = 4.12AEE464 pKa = 4.21EE465 pKa = 5.15EE466 pKa = 4.01YY467 pKa = 10.86DD468 pKa = 3.79GQSEE472 pKa = 4.21EE473 pKa = 4.17QYY475 pKa = 10.29EE476 pKa = 4.3AEE478 pKa = 4.89AEE480 pKa = 4.22PEE482 pKa = 4.05AQPEE486 pKa = 4.29DD487 pKa = 3.51QTEE490 pKa = 4.36TQLEE494 pKa = 4.39AEE496 pKa = 4.83PEE498 pKa = 4.49TPAGPLPIAEE508 pKa = 4.98DD509 pKa = 3.84KK510 pKa = 11.46DD511 pKa = 4.37NIWVSTGPHH520 pKa = 5.01TEE522 pKa = 3.46EE523 pKa = 4.69DD524 pKa = 3.71VAHH527 pKa = 7.06RR528 pKa = 11.84EE529 pKa = 4.16SAEE532 pKa = 4.11CTKK535 pKa = 11.02DD536 pKa = 3.37DD537 pKa = 3.75EE538 pKa = 4.52KK539 pKa = 11.62VYY541 pKa = 10.88GFIMKK546 pKa = 10.06HH547 pKa = 5.22NPSLLGDD554 pKa = 3.87AEE556 pKa = 4.36QPFRR560 pKa = 11.84CDD562 pKa = 4.68LYY564 pKa = 9.41NQCRR568 pKa = 11.84NTITHH573 pKa = 5.73VSIDD577 pKa = 3.35LSIYY581 pKa = 10.55EE582 pKa = 4.8KK583 pKa = 11.07GSLPNQEE590 pKa = 3.4VWKK593 pKa = 10.62SFKK596 pKa = 10.45NLQGFEE602 pKa = 3.76IAGNKK607 pKa = 7.1NTEE610 pKa = 4.01ALPNLSFALEE620 pKa = 4.17DD621 pKa = 3.37VSAAVSDD628 pKa = 4.4QEE630 pKa = 4.15DD631 pKa = 3.4QKK633 pKa = 11.46LSKK636 pKa = 10.57LVLSGVDD643 pKa = 3.67LSEE646 pKa = 4.1QTEE649 pKa = 4.07ALQNLKK655 pKa = 10.15NVRR658 pKa = 11.84KK659 pKa = 9.86LYY661 pKa = 10.95LFDD664 pKa = 3.5STFNASEE671 pKa = 4.1GAPNLLLTLFQCPGLSEE688 pKa = 4.6LKK690 pKa = 10.46VISSSISAIPSAGEE704 pKa = 3.74PASSCLSLHH713 pKa = 6.77ILRR716 pKa = 11.84CSQEE720 pKa = 4.07TVRR723 pKa = 11.84ALACTPASTFATVILNEE740 pKa = 4.08NTLGDD745 pKa = 4.38LEE747 pKa = 4.67CLNDD751 pKa = 3.96AEE753 pKa = 4.79SSIASLVIQSDD764 pKa = 3.61QTIKK768 pKa = 10.69SVKK771 pKa = 10.26KK772 pKa = 10.34SDD774 pKa = 3.82LQALPALKK782 pKa = 10.24SLCLFDD788 pKa = 5.16LQPSYY793 pKa = 11.05EE794 pKa = 4.39IDD796 pKa = 3.77PEE798 pKa = 4.06ILGPDD803 pKa = 3.55MPVSEE808 pKa = 4.81IFFSEE813 pKa = 3.9NEE815 pKa = 3.92EE816 pKa = 4.46FNFSSALPKK825 pKa = 10.26EE826 pKa = 4.17YY827 pKa = 10.67LKK829 pKa = 11.05NKK831 pKa = 9.14HH832 pKa = 6.2LGVYY836 pKa = 7.15THH838 pKa = 6.61GKK840 pKa = 9.0IEE842 pKa = 3.96NEE844 pKa = 4.28RR845 pKa = 11.84IFHH848 pKa = 6.4LVVDD852 pKa = 4.97PDD854 pKa = 3.69AKK856 pKa = 10.13TALLVIDD863 pKa = 4.92SPNDD867 pKa = 3.28FANTSEE873 pKa = 4.34AFKK876 pKa = 11.34KK877 pKa = 9.83MVEE880 pKa = 4.44SVNEE884 pKa = 3.87LDD886 pKa = 4.05TINVVVNTTDD896 pKa = 4.16DD897 pKa = 3.78SQKK900 pKa = 10.72EE901 pKa = 4.07AVTQKK906 pKa = 9.88ILSIFSHH913 pKa = 7.0LKK915 pKa = 7.0TQPTVDD921 pKa = 3.74YY922 pKa = 9.94TPKK925 pKa = 10.67NN926 pKa = 3.48
MM1 pKa = 7.68RR2 pKa = 11.84RR3 pKa = 11.84QAQHH7 pKa = 5.66RR8 pKa = 11.84CKK10 pKa = 10.59AILLLASLYY19 pKa = 9.31VLCGCIEE26 pKa = 4.18IDD28 pKa = 3.47PVVISTLEE36 pKa = 4.08DD37 pKa = 3.33FAKK40 pKa = 10.68DD41 pKa = 3.64LNEE44 pKa = 5.32KK45 pKa = 8.73PTIDD49 pKa = 3.09QNMFDD54 pKa = 4.47EE55 pKa = 5.26EE56 pKa = 4.27IKK58 pKa = 11.09ALTEE62 pKa = 3.92AKK64 pKa = 10.36LATEE68 pKa = 4.27EE69 pKa = 5.21KK70 pKa = 10.02IDD72 pKa = 3.87LLKK75 pKa = 10.77QRR77 pKa = 11.84GVQSGLATEE86 pKa = 4.6EE87 pKa = 4.48SNVPEE92 pKa = 4.19EE93 pKa = 3.92QSPAEE98 pKa = 4.03EE99 pKa = 3.76QLQAEE104 pKa = 4.57EE105 pKa = 4.06LAIGNDD111 pKa = 3.49EE112 pKa = 4.02QSEE115 pKa = 4.43VEE117 pKa = 4.17PEE119 pKa = 4.09VEE121 pKa = 4.05EE122 pKa = 4.36EE123 pKa = 3.97PTVFDD128 pKa = 3.64EE129 pKa = 4.01QSEE132 pKa = 4.55YY133 pKa = 11.06EE134 pKa = 4.09EE135 pKa = 4.41EE136 pKa = 4.07EE137 pKa = 4.19QYY139 pKa = 10.84EE140 pKa = 4.33AEE142 pKa = 4.52PATEE146 pKa = 3.74PTVFDD151 pKa = 3.66EE152 pKa = 4.29QLEE155 pKa = 4.35EE156 pKa = 4.22EE157 pKa = 4.87YY158 pKa = 10.69EE159 pKa = 4.32AEE161 pKa = 4.26PEE163 pKa = 4.06AADD166 pKa = 3.49EE167 pKa = 4.64AEE169 pKa = 4.11YY170 pKa = 10.9DD171 pKa = 3.88GQPEE175 pKa = 4.22EE176 pKa = 4.26QYY178 pKa = 10.53EE179 pKa = 4.12AQPEE183 pKa = 4.22PEE185 pKa = 4.25AEE187 pKa = 3.94EE188 pKa = 4.46EE189 pKa = 4.38YY190 pKa = 10.84EE191 pKa = 4.29PEE193 pKa = 4.48AEE195 pKa = 4.16PEE197 pKa = 3.97AEE199 pKa = 4.31EE200 pKa = 5.22EE201 pKa = 4.12YY202 pKa = 10.86DD203 pKa = 3.77GQPEE207 pKa = 4.09EE208 pKa = 4.21QYY210 pKa = 10.66EE211 pKa = 4.44AEE213 pKa = 4.46PEE215 pKa = 3.92AADD218 pKa = 4.44GIEE221 pKa = 3.79YY222 pKa = 10.43DD223 pKa = 3.92GQSEE227 pKa = 4.37EE228 pKa = 4.18QYY230 pKa = 10.2EE231 pKa = 4.24AQPEE235 pKa = 4.22AEE237 pKa = 5.05AEE239 pKa = 4.13AEE241 pKa = 4.21EE242 pKa = 4.15QSEE245 pKa = 4.39PEE247 pKa = 4.2EE248 pKa = 4.13QPEE251 pKa = 4.08AEE253 pKa = 4.17PEE255 pKa = 4.22VFDD258 pKa = 4.25EE259 pKa = 4.21QLEE262 pKa = 4.35AEE264 pKa = 4.66PEE266 pKa = 4.12PEE268 pKa = 4.17PEE270 pKa = 5.17AEE272 pKa = 4.37AEE274 pKa = 3.88YY275 pKa = 10.85DD276 pKa = 3.94GQSEE280 pKa = 4.61EE281 pKa = 3.98QDD283 pKa = 3.15EE284 pKa = 4.57AQPEE288 pKa = 4.44TQLEE292 pKa = 4.25AEE294 pKa = 4.43PEE296 pKa = 3.99AEE298 pKa = 4.22EE299 pKa = 5.15EE300 pKa = 4.01YY301 pKa = 10.86DD302 pKa = 3.79GQSEE306 pKa = 4.18EE307 pKa = 4.22QYY309 pKa = 10.84EE310 pKa = 4.38PEE312 pKa = 4.31AQPEE316 pKa = 4.17AEE318 pKa = 4.31PEE320 pKa = 3.96AEE322 pKa = 4.21AEE324 pKa = 4.48AEE326 pKa = 4.05AEE328 pKa = 3.84YY329 pKa = 10.87DD330 pKa = 3.83GQSEE334 pKa = 4.44EE335 pKa = 4.18QYY337 pKa = 9.98EE338 pKa = 4.34AQPEE342 pKa = 4.22TEE344 pKa = 3.76PAVFDD349 pKa = 3.96EE350 pKa = 4.26QLEE353 pKa = 4.34AEE355 pKa = 4.59PEE357 pKa = 4.11AEE359 pKa = 4.16PEE361 pKa = 4.08AEE363 pKa = 4.58PEE365 pKa = 4.49AEE367 pKa = 4.24TQLEE371 pKa = 4.24AEE373 pKa = 4.62PEE375 pKa = 4.06AEE377 pKa = 4.2AEE379 pKa = 4.48AEE381 pKa = 4.05AEE383 pKa = 3.84YY384 pKa = 10.87DD385 pKa = 3.83GQSEE389 pKa = 4.3EE390 pKa = 4.17QYY392 pKa = 10.24EE393 pKa = 4.25AEE395 pKa = 4.78PEE397 pKa = 4.14PEE399 pKa = 4.22TQLEE403 pKa = 4.34AEE405 pKa = 4.62PEE407 pKa = 4.09AEE409 pKa = 4.27PEE411 pKa = 4.2AEE413 pKa = 4.28ADD415 pKa = 3.37YY416 pKa = 11.23DD417 pKa = 3.97GQSEE421 pKa = 4.61YY422 pKa = 10.56EE423 pKa = 3.83AEE425 pKa = 4.0YY426 pKa = 10.9DD427 pKa = 4.57GEE429 pKa = 4.79SEE431 pKa = 4.26TQLEE435 pKa = 4.24EE436 pKa = 4.09EE437 pKa = 4.4EE438 pKa = 4.36QYY440 pKa = 10.85EE441 pKa = 4.37AEE443 pKa = 4.53PEE445 pKa = 3.96PTVYY449 pKa = 10.62DD450 pKa = 3.82EE451 pKa = 4.23QLEE454 pKa = 4.37AEE456 pKa = 4.58PEE458 pKa = 4.07AEE460 pKa = 3.97PEE462 pKa = 4.12AEE464 pKa = 4.21EE465 pKa = 5.15EE466 pKa = 4.01YY467 pKa = 10.86DD468 pKa = 3.79GQSEE472 pKa = 4.21EE473 pKa = 4.17QYY475 pKa = 10.29EE476 pKa = 4.3AEE478 pKa = 4.89AEE480 pKa = 4.22PEE482 pKa = 4.05AQPEE486 pKa = 4.29DD487 pKa = 3.51QTEE490 pKa = 4.36TQLEE494 pKa = 4.39AEE496 pKa = 4.83PEE498 pKa = 4.49TPAGPLPIAEE508 pKa = 4.98DD509 pKa = 3.84KK510 pKa = 11.46DD511 pKa = 4.37NIWVSTGPHH520 pKa = 5.01TEE522 pKa = 3.46EE523 pKa = 4.69DD524 pKa = 3.71VAHH527 pKa = 7.06RR528 pKa = 11.84EE529 pKa = 4.16SAEE532 pKa = 4.11CTKK535 pKa = 11.02DD536 pKa = 3.37DD537 pKa = 3.75EE538 pKa = 4.52KK539 pKa = 11.62VYY541 pKa = 10.88GFIMKK546 pKa = 10.06HH547 pKa = 5.22NPSLLGDD554 pKa = 3.87AEE556 pKa = 4.36QPFRR560 pKa = 11.84CDD562 pKa = 4.68LYY564 pKa = 9.41NQCRR568 pKa = 11.84NTITHH573 pKa = 5.73VSIDD577 pKa = 3.35LSIYY581 pKa = 10.55EE582 pKa = 4.8KK583 pKa = 11.07GSLPNQEE590 pKa = 3.4VWKK593 pKa = 10.62SFKK596 pKa = 10.45NLQGFEE602 pKa = 3.76IAGNKK607 pKa = 7.1NTEE610 pKa = 4.01ALPNLSFALEE620 pKa = 4.17DD621 pKa = 3.37VSAAVSDD628 pKa = 4.4QEE630 pKa = 4.15DD631 pKa = 3.4QKK633 pKa = 11.46LSKK636 pKa = 10.57LVLSGVDD643 pKa = 3.67LSEE646 pKa = 4.1QTEE649 pKa = 4.07ALQNLKK655 pKa = 10.15NVRR658 pKa = 11.84KK659 pKa = 9.86LYY661 pKa = 10.95LFDD664 pKa = 3.5STFNASEE671 pKa = 4.1GAPNLLLTLFQCPGLSEE688 pKa = 4.6LKK690 pKa = 10.46VISSSISAIPSAGEE704 pKa = 3.74PASSCLSLHH713 pKa = 6.77ILRR716 pKa = 11.84CSQEE720 pKa = 4.07TVRR723 pKa = 11.84ALACTPASTFATVILNEE740 pKa = 4.08NTLGDD745 pKa = 4.38LEE747 pKa = 4.67CLNDD751 pKa = 3.96AEE753 pKa = 4.79SSIASLVIQSDD764 pKa = 3.61QTIKK768 pKa = 10.69SVKK771 pKa = 10.26KK772 pKa = 10.34SDD774 pKa = 3.82LQALPALKK782 pKa = 10.24SLCLFDD788 pKa = 5.16LQPSYY793 pKa = 11.05EE794 pKa = 4.39IDD796 pKa = 3.77PEE798 pKa = 4.06ILGPDD803 pKa = 3.55MPVSEE808 pKa = 4.81IFFSEE813 pKa = 3.9NEE815 pKa = 3.92EE816 pKa = 4.46FNFSSALPKK825 pKa = 10.26EE826 pKa = 4.17YY827 pKa = 10.67LKK829 pKa = 11.05NKK831 pKa = 9.14HH832 pKa = 6.2LGVYY836 pKa = 7.15THH838 pKa = 6.61GKK840 pKa = 9.0IEE842 pKa = 3.96NEE844 pKa = 4.28RR845 pKa = 11.84IFHH848 pKa = 6.4LVVDD852 pKa = 4.97PDD854 pKa = 3.69AKK856 pKa = 10.13TALLVIDD863 pKa = 4.92SPNDD867 pKa = 3.28FANTSEE873 pKa = 4.34AFKK876 pKa = 11.34KK877 pKa = 9.83MVEE880 pKa = 4.44SVNEE884 pKa = 3.87LDD886 pKa = 4.05TINVVVNTTDD896 pKa = 4.16DD897 pKa = 3.78SQKK900 pKa = 10.72EE901 pKa = 4.07AVTQKK906 pKa = 9.88ILSIFSHH913 pKa = 7.0LKK915 pKa = 7.0TQPTVDD921 pKa = 3.74YY922 pKa = 9.94TPKK925 pKa = 10.67NN926 pKa = 3.48
Molecular weight: 103.56 kDa
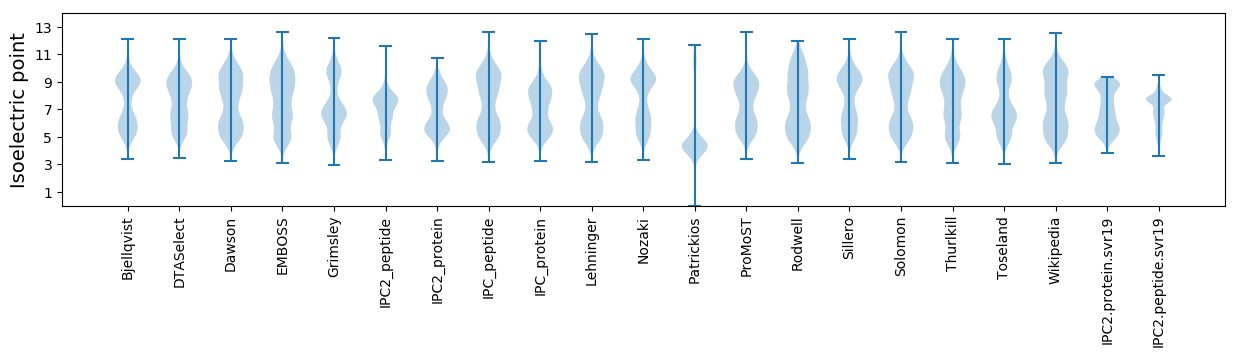
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177EJP7|A0A177EJP7_9MICR Serine/threonine-protein kinase MRCK OS=Nematocida displodere OX=1805483 GN=NEDG_01622 PE=4 SV=1
MM1 pKa = 7.08IWGIEE6 pKa = 3.96RR7 pKa = 11.84LSVHH11 pKa = 5.81RR12 pKa = 11.84QKK14 pKa = 11.16EE15 pKa = 4.23WTHH18 pKa = 5.03TLAAPFLKK26 pKa = 9.65LTASLTRR33 pKa = 11.84THH35 pKa = 6.56VIISATYY42 pKa = 8.23FGSEE46 pKa = 3.97EE47 pKa = 4.06VLVCVLAAAATSVHH61 pKa = 6.22CSKK64 pKa = 11.05DD65 pKa = 3.22LALFAGPLRR74 pKa = 11.84VQLKK78 pKa = 9.7LCSARR83 pKa = 11.84AAQTLHH89 pKa = 6.51RR90 pKa = 11.84RR91 pKa = 11.84LIYY94 pKa = 6.91TQKK97 pKa = 9.5MLRR100 pKa = 11.84FIQGMQKK107 pKa = 9.76VRR109 pKa = 11.84LPSASAPGPPRR120 pKa = 4.47
MM1 pKa = 7.08IWGIEE6 pKa = 3.96RR7 pKa = 11.84LSVHH11 pKa = 5.81RR12 pKa = 11.84QKK14 pKa = 11.16EE15 pKa = 4.23WTHH18 pKa = 5.03TLAAPFLKK26 pKa = 9.65LTASLTRR33 pKa = 11.84THH35 pKa = 6.56VIISATYY42 pKa = 8.23FGSEE46 pKa = 3.97EE47 pKa = 4.06VLVCVLAAAATSVHH61 pKa = 6.22CSKK64 pKa = 11.05DD65 pKa = 3.22LALFAGPLRR74 pKa = 11.84VQLKK78 pKa = 9.7LCSARR83 pKa = 11.84AAQTLHH89 pKa = 6.51RR90 pKa = 11.84RR91 pKa = 11.84LIYY94 pKa = 6.91TQKK97 pKa = 9.5MLRR100 pKa = 11.84FIQGMQKK107 pKa = 9.76VRR109 pKa = 11.84LPSASAPGPPRR120 pKa = 4.47
Molecular weight: 13.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
862370 |
52 |
3243 |
384.8 |
43.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.04 ± 0.047 | 1.76 ± 0.024 |
4.327 ± 0.03 | 7.322 ± 0.062 |
4.13 ± 0.036 | 5.61 ± 0.044 |
2.278 ± 0.02 | 6.396 ± 0.036 |
6.806 ± 0.048 | 10.434 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.24 ± 0.019 | 3.686 ± 0.037 |
4.688 ± 0.053 | 3.469 ± 0.027 |
5.04 ± 0.034 | 7.65 ± 0.047 |
6.499 ± 0.041 | 6.659 ± 0.04 |
0.869 ± 0.014 | 3.096 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
