
Rodent Torque teno virus 1
Taxonomy: Viruses; Anelloviridae; Rhotorquevirus; Torque teno rodent virus 1
Average proteome isoelectric point is 7.88
Get precalculated fractions of proteins
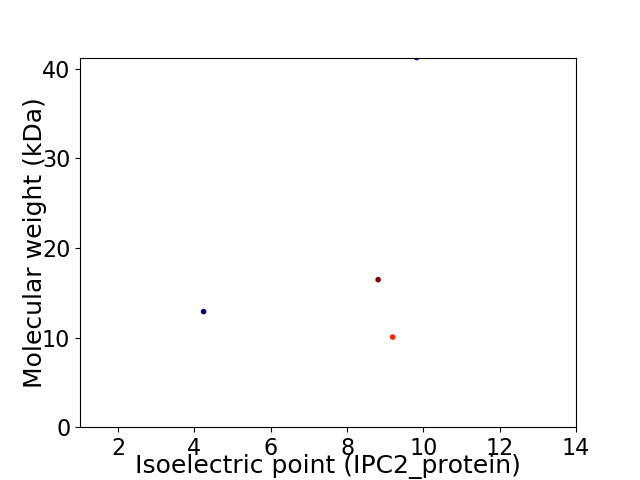
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
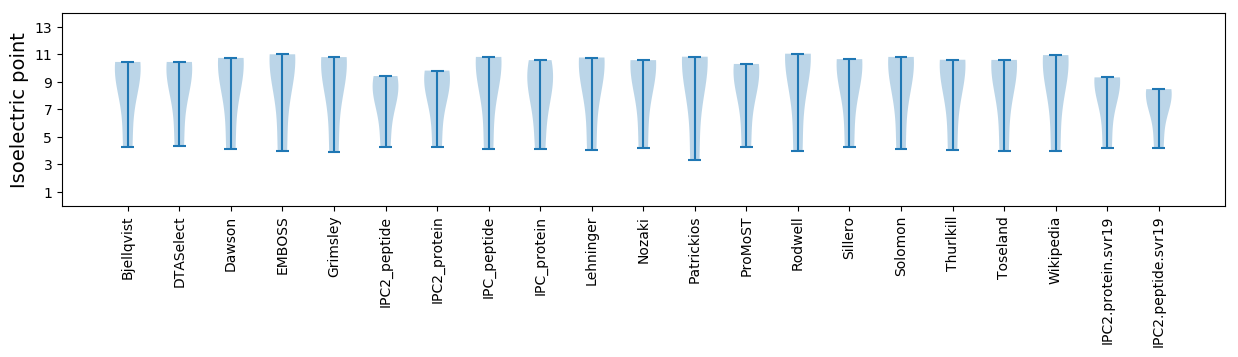
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X2G8B9|X2G8B9_9VIRU ORF3 OS=Rodent Torque teno virus 1 OX=1514664 PE=4 SV=1
MM1 pKa = 7.23NVDD4 pKa = 2.66EE5 pKa = 4.83DD6 pKa = 4.48FIYY9 pKa = 10.85RR10 pKa = 11.84EE11 pKa = 4.12DD12 pKa = 3.36SWLCMIKK19 pKa = 8.47QTHH22 pKa = 5.99EE23 pKa = 4.17LFCNCTSWTKK33 pKa = 10.31HH34 pKa = 5.57LKK36 pKa = 8.96TLLDD40 pKa = 3.97FNHH43 pKa = 6.94PEE45 pKa = 3.84WRR47 pKa = 11.84GDD49 pKa = 3.79SQDD52 pKa = 3.6EE53 pKa = 4.23QPGKK57 pKa = 10.26EE58 pKa = 4.01KK59 pKa = 10.76PGDD62 pKa = 3.7NPEE65 pKa = 4.21GVPTDD70 pKa = 3.68EE71 pKa = 4.4EE72 pKa = 4.91LIGALEE78 pKa = 3.84EE79 pKa = 4.61LEE81 pKa = 4.18NGGAGEE87 pKa = 4.21GTSRR91 pKa = 11.84SEE93 pKa = 4.09CSLKK97 pKa = 10.9YY98 pKa = 10.87SLDD101 pKa = 3.45TLGDD105 pKa = 3.73VLSEE109 pKa = 4.07ALCVLSS115 pKa = 4.57
MM1 pKa = 7.23NVDD4 pKa = 2.66EE5 pKa = 4.83DD6 pKa = 4.48FIYY9 pKa = 10.85RR10 pKa = 11.84EE11 pKa = 4.12DD12 pKa = 3.36SWLCMIKK19 pKa = 8.47QTHH22 pKa = 5.99EE23 pKa = 4.17LFCNCTSWTKK33 pKa = 10.31HH34 pKa = 5.57LKK36 pKa = 8.96TLLDD40 pKa = 3.97FNHH43 pKa = 6.94PEE45 pKa = 3.84WRR47 pKa = 11.84GDD49 pKa = 3.79SQDD52 pKa = 3.6EE53 pKa = 4.23QPGKK57 pKa = 10.26EE58 pKa = 4.01KK59 pKa = 10.76PGDD62 pKa = 3.7NPEE65 pKa = 4.21GVPTDD70 pKa = 3.68EE71 pKa = 4.4EE72 pKa = 4.91LIGALEE78 pKa = 3.84EE79 pKa = 4.61LEE81 pKa = 4.18NGGAGEE87 pKa = 4.21GTSRR91 pKa = 11.84SEE93 pKa = 4.09CSLKK97 pKa = 10.9YY98 pKa = 10.87SLDD101 pKa = 3.45TLGDD105 pKa = 3.73VLSEE109 pKa = 4.07ALCVLSS115 pKa = 4.57
Molecular weight: 12.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X2G9U7|X2G9U7_9VIRU Capsid protein OS=Rodent Torque teno virus 1 OX=1514664 PE=3 SV=1
MM1 pKa = 7.63AWRR4 pKa = 11.84FPRR7 pKa = 11.84RR8 pKa = 11.84TTWKK12 pKa = 9.66RR13 pKa = 11.84KK14 pKa = 5.77TWRR17 pKa = 11.84QPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84TYY24 pKa = 10.0RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84AYY29 pKa = 10.18RR30 pKa = 11.84RR31 pKa = 11.84TRR33 pKa = 11.84RR34 pKa = 11.84TRR36 pKa = 11.84KK37 pKa = 7.06WRR39 pKa = 11.84RR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84NLKK46 pKa = 9.82VRR48 pKa = 11.84VLTEE52 pKa = 3.35IQPRR56 pKa = 11.84YY57 pKa = 7.46TRR59 pKa = 11.84RR60 pKa = 11.84CVIRR64 pKa = 11.84GIMCPLIVGQPNADD78 pKa = 3.34GSPSKK83 pKa = 10.8QNWEE87 pKa = 3.96LRR89 pKa = 11.84QGGIFAYY96 pKa = 9.74KK97 pKa = 10.08HH98 pKa = 6.53DD99 pKa = 5.29GSDD102 pKa = 2.78LWMGTWSMGMVSLSILYY119 pKa = 10.29GEE121 pKa = 4.23HH122 pKa = 4.63QAYY125 pKa = 9.52RR126 pKa = 11.84NRR128 pKa = 11.84WSVSNCGFDD137 pKa = 3.15MVEE140 pKa = 3.87YY141 pKa = 10.35RR142 pKa = 11.84GTTIYY147 pKa = 10.8LEE149 pKa = 4.08QHH151 pKa = 5.0STMDD155 pKa = 3.65YY156 pKa = 10.57IVFFDD161 pKa = 3.66EE162 pKa = 4.72EE163 pKa = 4.41YY164 pKa = 11.15RR165 pKa = 11.84DD166 pKa = 3.53ITSFAAQASLHH177 pKa = 6.2PLVLMTHH184 pKa = 7.4PKK186 pKa = 9.56TILIKK191 pKa = 9.96SRR193 pKa = 11.84EE194 pKa = 3.98RR195 pKa = 11.84AGPRR199 pKa = 11.84RR200 pKa = 11.84ARR202 pKa = 11.84KK203 pKa = 9.73VFIPRR208 pKa = 11.84PSWWDD213 pKa = 3.44SGWKK217 pKa = 9.58FSKK220 pKa = 10.53DD221 pKa = 3.46VCNKK225 pKa = 10.26GLFVWYY231 pKa = 9.85ISAIDD236 pKa = 4.11MEE238 pKa = 4.98HH239 pKa = 6.9PWMKK243 pKa = 10.57AYY245 pKa = 10.44VGNKK249 pKa = 8.57TDD251 pKa = 4.03FQNLMWWRR259 pKa = 11.84NNKK262 pKa = 8.4WKK264 pKa = 10.54EE265 pKa = 3.96DD266 pKa = 2.96FDD268 pKa = 4.27TYY270 pKa = 11.55VRR272 pKa = 11.84TTAQMLQTSKK282 pKa = 11.22NDD284 pKa = 3.31NNYY287 pKa = 8.53KK288 pKa = 9.64TEE290 pKa = 4.06TATGPFMLSIKK301 pKa = 10.09DD302 pKa = 3.62RR303 pKa = 11.84NRR305 pKa = 11.84ATVNQQFTWFYY316 pKa = 10.06KK317 pKa = 10.54SYY319 pKa = 9.11WHH321 pKa = 7.09WGGNNLTIKK330 pKa = 10.21KK331 pKa = 9.35VCNPNMDD338 pKa = 3.43VPITT342 pKa = 3.68
MM1 pKa = 7.63AWRR4 pKa = 11.84FPRR7 pKa = 11.84RR8 pKa = 11.84TTWKK12 pKa = 9.66RR13 pKa = 11.84KK14 pKa = 5.77TWRR17 pKa = 11.84QPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84TYY24 pKa = 10.0RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84AYY29 pKa = 10.18RR30 pKa = 11.84RR31 pKa = 11.84TRR33 pKa = 11.84RR34 pKa = 11.84TRR36 pKa = 11.84KK37 pKa = 7.06WRR39 pKa = 11.84RR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84NLKK46 pKa = 9.82VRR48 pKa = 11.84VLTEE52 pKa = 3.35IQPRR56 pKa = 11.84YY57 pKa = 7.46TRR59 pKa = 11.84RR60 pKa = 11.84CVIRR64 pKa = 11.84GIMCPLIVGQPNADD78 pKa = 3.34GSPSKK83 pKa = 10.8QNWEE87 pKa = 3.96LRR89 pKa = 11.84QGGIFAYY96 pKa = 9.74KK97 pKa = 10.08HH98 pKa = 6.53DD99 pKa = 5.29GSDD102 pKa = 2.78LWMGTWSMGMVSLSILYY119 pKa = 10.29GEE121 pKa = 4.23HH122 pKa = 4.63QAYY125 pKa = 9.52RR126 pKa = 11.84NRR128 pKa = 11.84WSVSNCGFDD137 pKa = 3.15MVEE140 pKa = 3.87YY141 pKa = 10.35RR142 pKa = 11.84GTTIYY147 pKa = 10.8LEE149 pKa = 4.08QHH151 pKa = 5.0STMDD155 pKa = 3.65YY156 pKa = 10.57IVFFDD161 pKa = 3.66EE162 pKa = 4.72EE163 pKa = 4.41YY164 pKa = 11.15RR165 pKa = 11.84DD166 pKa = 3.53ITSFAAQASLHH177 pKa = 6.2PLVLMTHH184 pKa = 7.4PKK186 pKa = 9.56TILIKK191 pKa = 9.96SRR193 pKa = 11.84EE194 pKa = 3.98RR195 pKa = 11.84AGPRR199 pKa = 11.84RR200 pKa = 11.84ARR202 pKa = 11.84KK203 pKa = 9.73VFIPRR208 pKa = 11.84PSWWDD213 pKa = 3.44SGWKK217 pKa = 9.58FSKK220 pKa = 10.53DD221 pKa = 3.46VCNKK225 pKa = 10.26GLFVWYY231 pKa = 9.85ISAIDD236 pKa = 4.11MEE238 pKa = 4.98HH239 pKa = 6.9PWMKK243 pKa = 10.57AYY245 pKa = 10.44VGNKK249 pKa = 8.57TDD251 pKa = 4.03FQNLMWWRR259 pKa = 11.84NNKK262 pKa = 8.4WKK264 pKa = 10.54EE265 pKa = 3.96DD266 pKa = 2.96FDD268 pKa = 4.27TYY270 pKa = 11.55VRR272 pKa = 11.84TTAQMLQTSKK282 pKa = 11.22NDD284 pKa = 3.31NNYY287 pKa = 8.53KK288 pKa = 9.64TEE290 pKa = 4.06TATGPFMLSIKK301 pKa = 10.09DD302 pKa = 3.62RR303 pKa = 11.84NRR305 pKa = 11.84ATVNQQFTWFYY316 pKa = 10.06KK317 pKa = 10.54SYY319 pKa = 9.11WHH321 pKa = 7.09WGGNNLTIKK330 pKa = 10.21KK331 pKa = 9.35VCNPNMDD338 pKa = 3.43VPITT342 pKa = 3.68
Molecular weight: 41.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
685 |
83 |
342 |
171.3 |
20.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.65 ± 0.927 | 2.19 ± 0.595 |
5.109 ± 0.82 | 6.277 ± 2.202 |
3.358 ± 0.451 | 5.985 ± 0.758 |
2.336 ± 0.424 | 4.088 ± 0.917 |
7.591 ± 1.039 | 6.861 ± 1.593 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.212 ± 0.574 | 4.964 ± 0.378 |
4.818 ± 0.398 | 4.818 ± 0.916 |
8.905 ± 2.127 | 7.153 ± 1.907 |
7.153 ± 0.699 | 4.38 ± 0.555 |
4.234 ± 0.839 | 2.92 ± 1.067 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
