
Pelagirhabdus alkalitolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Pelagirhabdus
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
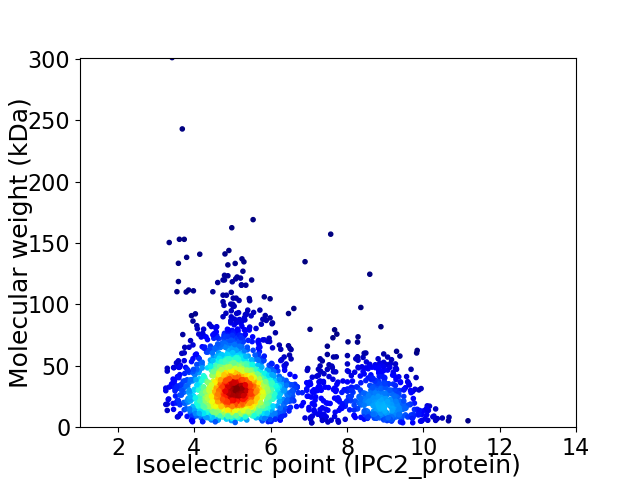
Virtual 2D-PAGE plot for 2406 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6GQV9|A0A1G6GQV9_9BACI Histidine kinase OS=Pelagirhabdus alkalitolerans OX=1612202 GN=SAMN05421734_101408 PE=4 SV=1
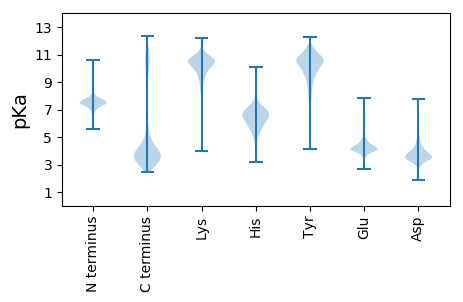
MM1 pKa = 6.69MKK3 pKa = 10.17KK4 pKa = 9.77YY5 pKa = 11.0VKK7 pKa = 10.48GFLFAVLITLVLAGCGNGDD26 pKa = 4.12EE27 pKa = 4.92NGNDD31 pKa = 3.29QAGDD35 pKa = 3.83SEE37 pKa = 4.75AEE39 pKa = 4.12AVEE42 pKa = 4.15LTLFSSVTNEE52 pKa = 3.2SDD54 pKa = 3.2QAIFEE59 pKa = 4.27EE60 pKa = 4.73VIEE63 pKa = 4.31RR64 pKa = 11.84FEE66 pKa = 4.25EE67 pKa = 4.37EE68 pKa = 3.99NDD70 pKa = 4.37HH71 pKa = 7.14IQIDD75 pKa = 4.37YY76 pKa = 10.77NFPTSEE82 pKa = 4.43YY83 pKa = 8.69EE84 pKa = 3.75SMMRR88 pKa = 11.84VRR90 pKa = 11.84MGADD94 pKa = 3.33DD95 pKa = 4.34MPDD98 pKa = 3.16LFDD101 pKa = 3.36THH103 pKa = 5.67GWAVNRR109 pKa = 11.84YY110 pKa = 8.83GDD112 pKa = 3.62YY113 pKa = 11.34TMDD116 pKa = 4.17LRR118 pKa = 11.84DD119 pKa = 4.15MDD121 pKa = 4.0WVDD124 pKa = 4.07NLDD127 pKa = 4.0PSLEE131 pKa = 4.34PIITDD136 pKa = 4.06DD137 pKa = 3.95DD138 pKa = 4.69GKK140 pKa = 11.41VYY142 pKa = 10.32AYY144 pKa = 10.06PMNQANDD151 pKa = 3.55GLTYY155 pKa = 10.76NVGILEE161 pKa = 4.74EE162 pKa = 4.16YY163 pKa = 10.63DD164 pKa = 2.97IDD166 pKa = 4.01VPEE169 pKa = 4.59TFDD172 pKa = 3.78EE173 pKa = 4.93LMDD176 pKa = 4.26ALRR179 pKa = 11.84EE180 pKa = 4.09IRR182 pKa = 11.84DD183 pKa = 3.52QSDD186 pKa = 4.08GEE188 pKa = 4.44VTPLWFPGSDD198 pKa = 3.37RR199 pKa = 11.84YY200 pKa = 11.12SLGYY204 pKa = 10.21FLDD207 pKa = 3.68IMSTPLLVTHH217 pKa = 6.97EE218 pKa = 4.1DD219 pKa = 3.06HH220 pKa = 8.04DD221 pKa = 4.58YY222 pKa = 11.54SEE224 pKa = 4.26EE225 pKa = 4.07LKK227 pKa = 10.9AGEE230 pKa = 4.44YY231 pKa = 10.73GWDD234 pKa = 3.41EE235 pKa = 4.0YY236 pKa = 11.42TYY238 pKa = 11.07LPEE241 pKa = 6.2KK242 pKa = 9.57LLQMQEE248 pKa = 3.92EE249 pKa = 4.49DD250 pKa = 4.03LLNVDD255 pKa = 3.77VLTAQTHH262 pKa = 5.11EE263 pKa = 4.18QTQLFAQDD271 pKa = 3.32KK272 pKa = 9.43IGFVMGTLPVDD283 pKa = 4.65SIHH286 pKa = 6.61EE287 pKa = 4.16LNEE290 pKa = 4.74DD291 pKa = 3.44IEE293 pKa = 4.69LGIMPVPPIHH303 pKa = 7.81DD304 pKa = 4.15GGEE307 pKa = 4.01KK308 pKa = 9.95SWIGGEE314 pKa = 4.05RR315 pKa = 11.84FTLAVAQNSDD325 pKa = 3.36HH326 pKa = 7.05PEE328 pKa = 3.63EE329 pKa = 4.26AKK331 pKa = 10.71QFIEE335 pKa = 5.72FLAQPDD341 pKa = 3.54IAQKK345 pKa = 10.35VAEE348 pKa = 4.46GTNLPSGLEE357 pKa = 3.97NVSAEE362 pKa = 3.78IYY364 pKa = 9.87YY365 pKa = 9.84QDD367 pKa = 4.19FYY369 pKa = 11.88EE370 pKa = 5.0EE371 pKa = 4.25YY372 pKa = 10.42DD373 pKa = 3.77DD374 pKa = 6.16VIVEE378 pKa = 4.29PYY380 pKa = 9.85FDD382 pKa = 5.48RR383 pKa = 11.84EE384 pKa = 4.07FLPSGMWDD392 pKa = 3.4VYY394 pKa = 11.55GEE396 pKa = 4.21TAQEE400 pKa = 3.97LLSGRR405 pKa = 11.84MTPEE409 pKa = 3.4EE410 pKa = 4.16VTEE413 pKa = 4.24QMAAEE418 pKa = 4.1YY419 pKa = 10.62NRR421 pKa = 11.84LYY423 pKa = 10.98DD424 pKa = 3.78EE425 pKa = 4.47EE426 pKa = 4.33
MM1 pKa = 6.69MKK3 pKa = 10.17KK4 pKa = 9.77YY5 pKa = 11.0VKK7 pKa = 10.48GFLFAVLITLVLAGCGNGDD26 pKa = 4.12EE27 pKa = 4.92NGNDD31 pKa = 3.29QAGDD35 pKa = 3.83SEE37 pKa = 4.75AEE39 pKa = 4.12AVEE42 pKa = 4.15LTLFSSVTNEE52 pKa = 3.2SDD54 pKa = 3.2QAIFEE59 pKa = 4.27EE60 pKa = 4.73VIEE63 pKa = 4.31RR64 pKa = 11.84FEE66 pKa = 4.25EE67 pKa = 4.37EE68 pKa = 3.99NDD70 pKa = 4.37HH71 pKa = 7.14IQIDD75 pKa = 4.37YY76 pKa = 10.77NFPTSEE82 pKa = 4.43YY83 pKa = 8.69EE84 pKa = 3.75SMMRR88 pKa = 11.84VRR90 pKa = 11.84MGADD94 pKa = 3.33DD95 pKa = 4.34MPDD98 pKa = 3.16LFDD101 pKa = 3.36THH103 pKa = 5.67GWAVNRR109 pKa = 11.84YY110 pKa = 8.83GDD112 pKa = 3.62YY113 pKa = 11.34TMDD116 pKa = 4.17LRR118 pKa = 11.84DD119 pKa = 4.15MDD121 pKa = 4.0WVDD124 pKa = 4.07NLDD127 pKa = 4.0PSLEE131 pKa = 4.34PIITDD136 pKa = 4.06DD137 pKa = 3.95DD138 pKa = 4.69GKK140 pKa = 11.41VYY142 pKa = 10.32AYY144 pKa = 10.06PMNQANDD151 pKa = 3.55GLTYY155 pKa = 10.76NVGILEE161 pKa = 4.74EE162 pKa = 4.16YY163 pKa = 10.63DD164 pKa = 2.97IDD166 pKa = 4.01VPEE169 pKa = 4.59TFDD172 pKa = 3.78EE173 pKa = 4.93LMDD176 pKa = 4.26ALRR179 pKa = 11.84EE180 pKa = 4.09IRR182 pKa = 11.84DD183 pKa = 3.52QSDD186 pKa = 4.08GEE188 pKa = 4.44VTPLWFPGSDD198 pKa = 3.37RR199 pKa = 11.84YY200 pKa = 11.12SLGYY204 pKa = 10.21FLDD207 pKa = 3.68IMSTPLLVTHH217 pKa = 6.97EE218 pKa = 4.1DD219 pKa = 3.06HH220 pKa = 8.04DD221 pKa = 4.58YY222 pKa = 11.54SEE224 pKa = 4.26EE225 pKa = 4.07LKK227 pKa = 10.9AGEE230 pKa = 4.44YY231 pKa = 10.73GWDD234 pKa = 3.41EE235 pKa = 4.0YY236 pKa = 11.42TYY238 pKa = 11.07LPEE241 pKa = 6.2KK242 pKa = 9.57LLQMQEE248 pKa = 3.92EE249 pKa = 4.49DD250 pKa = 4.03LLNVDD255 pKa = 3.77VLTAQTHH262 pKa = 5.11EE263 pKa = 4.18QTQLFAQDD271 pKa = 3.32KK272 pKa = 9.43IGFVMGTLPVDD283 pKa = 4.65SIHH286 pKa = 6.61EE287 pKa = 4.16LNEE290 pKa = 4.74DD291 pKa = 3.44IEE293 pKa = 4.69LGIMPVPPIHH303 pKa = 7.81DD304 pKa = 4.15GGEE307 pKa = 4.01KK308 pKa = 9.95SWIGGEE314 pKa = 4.05RR315 pKa = 11.84FTLAVAQNSDD325 pKa = 3.36HH326 pKa = 7.05PEE328 pKa = 3.63EE329 pKa = 4.26AKK331 pKa = 10.71QFIEE335 pKa = 5.72FLAQPDD341 pKa = 3.54IAQKK345 pKa = 10.35VAEE348 pKa = 4.46GTNLPSGLEE357 pKa = 3.97NVSAEE362 pKa = 3.78IYY364 pKa = 9.87YY365 pKa = 9.84QDD367 pKa = 4.19FYY369 pKa = 11.88EE370 pKa = 5.0EE371 pKa = 4.25YY372 pKa = 10.42DD373 pKa = 3.77DD374 pKa = 6.16VIVEE378 pKa = 4.29PYY380 pKa = 9.85FDD382 pKa = 5.48RR383 pKa = 11.84EE384 pKa = 4.07FLPSGMWDD392 pKa = 3.4VYY394 pKa = 11.55GEE396 pKa = 4.21TAQEE400 pKa = 3.97LLSGRR405 pKa = 11.84MTPEE409 pKa = 3.4EE410 pKa = 4.16VTEE413 pKa = 4.24QMAAEE418 pKa = 4.1YY419 pKa = 10.62NRR421 pKa = 11.84LYY423 pKa = 10.98DD424 pKa = 3.78EE425 pKa = 4.47EE426 pKa = 4.33
Molecular weight: 48.7 kDa
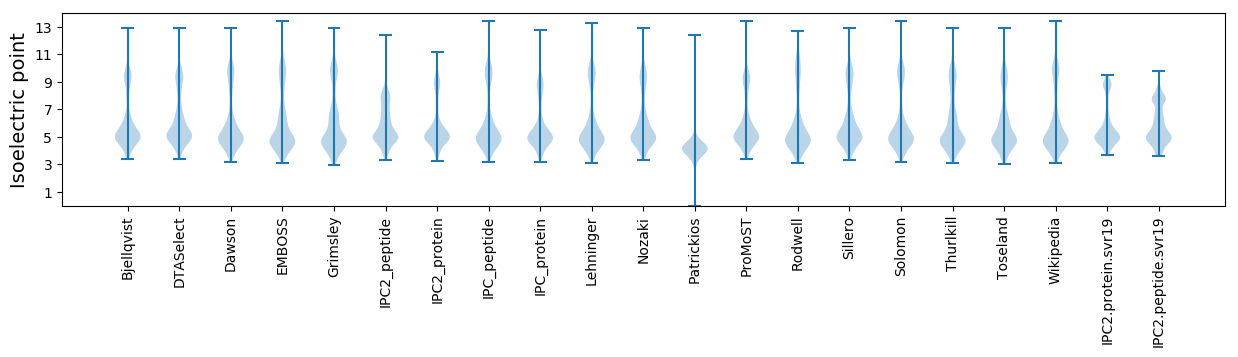
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6LCH0|A0A1G6LCH0_9BACI Uncharacterized protein OS=Pelagirhabdus alkalitolerans OX=1612202 GN=SAMN05421734_10835 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 8.86HH12 pKa = 6.39KK13 pKa = 9.52KK14 pKa = 7.68VHH16 pKa = 5.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSKK25 pKa = 10.35NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.36KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 8.86HH12 pKa = 6.39KK13 pKa = 9.52KK14 pKa = 7.68VHH16 pKa = 5.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSKK25 pKa = 10.35NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.36KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
744324 |
31 |
2683 |
309.4 |
34.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.419 ± 0.048 | 0.55 ± 0.012 |
6.432 ± 0.054 | 7.779 ± 0.066 |
4.375 ± 0.042 | 6.273 ± 0.049 |
2.359 ± 0.029 | 7.811 ± 0.05 |
6.088 ± 0.06 | 9.662 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.746 ± 0.025 | 4.411 ± 0.034 |
3.397 ± 0.028 | 4.295 ± 0.032 |
4.02 ± 0.032 | 6.155 ± 0.035 |
5.659 ± 0.034 | 7.024 ± 0.039 |
0.888 ± 0.017 | 3.656 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
