
Lake Sarah-associated circular virus-29
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.57
Get precalculated fractions of proteins
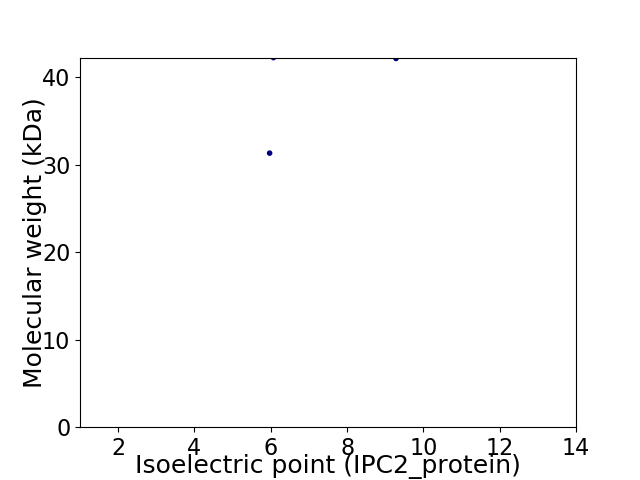
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA95|A0A126GA95_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-29 OX=1685756 PE=4 SV=1
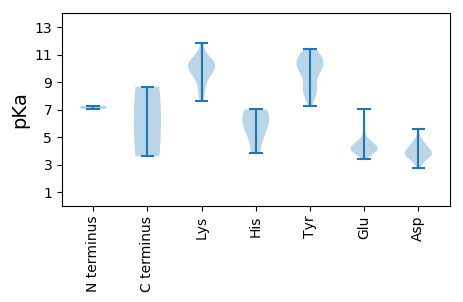
MM1 pKa = 7.27SVSDD5 pKa = 4.25DD6 pKa = 3.97FEE8 pKa = 7.01AGAEE12 pKa = 4.13PIQLYY17 pKa = 10.12GALLEE22 pKa = 4.19ICEE25 pKa = 4.12IFAYY29 pKa = 8.25QHH31 pKa = 5.75EE32 pKa = 4.68VAPTTGYY39 pKa = 10.22RR40 pKa = 11.84HH41 pKa = 4.6YY42 pKa = 10.61QGYY45 pKa = 10.34LEE47 pKa = 5.49LVTKK51 pKa = 10.49NRR53 pKa = 11.84HH54 pKa = 3.83TWIQNQLLKK63 pKa = 10.78HH64 pKa = 5.73GLHH67 pKa = 6.47FEE69 pKa = 4.28YY70 pKa = 10.79LVGAKK75 pKa = 9.76GLPKK79 pKa = 10.05QAWAYY84 pKa = 7.26ATKK87 pKa = 10.36EE88 pKa = 4.1DD89 pKa = 4.14TRR91 pKa = 11.84ADD93 pKa = 4.04GPWTFGEE100 pKa = 4.26PRR102 pKa = 11.84DD103 pKa = 3.81SEE105 pKa = 4.36KK106 pKa = 11.27ANKK109 pKa = 9.53SWLFVKK115 pKa = 10.38AIKK118 pKa = 10.52SGATDD123 pKa = 4.67AEE125 pKa = 4.54LCDD128 pKa = 4.31SFPGMMTNSFSNANRR143 pKa = 11.84IRR145 pKa = 11.84QAYY148 pKa = 8.11GVQLPEE154 pKa = 4.19PEE156 pKa = 3.83RR157 pKa = 11.84TVPLEE162 pKa = 3.96LYY164 pKa = 10.48LFYY167 pKa = 11.02GPSGTGKK174 pKa = 9.76SQFARR179 pKa = 11.84DD180 pKa = 3.51QARR183 pKa = 11.84LAKK186 pKa = 9.17MNPYY190 pKa = 9.71VLPIGKK196 pKa = 9.33DD197 pKa = 3.47FWVTPAMCGKK207 pKa = 9.94KK208 pKa = 10.47YY209 pKa = 10.95YY210 pKa = 10.19IIDD213 pKa = 3.98DD214 pKa = 4.26FKK216 pKa = 11.85SNLGLSDD223 pKa = 4.69LLRR226 pKa = 11.84LLDD229 pKa = 4.15TYY231 pKa = 10.9PVEE234 pKa = 4.6APNKK238 pKa = 9.42GIASSEE244 pKa = 3.89FSSLNGGVFNNFVRR258 pKa = 11.84WPYY261 pKa = 10.87VVDD264 pKa = 3.21ARR266 pKa = 11.84NYY268 pKa = 8.35CHH270 pKa = 6.75YY271 pKa = 9.77NQSITSS277 pKa = 3.6
MM1 pKa = 7.27SVSDD5 pKa = 4.25DD6 pKa = 3.97FEE8 pKa = 7.01AGAEE12 pKa = 4.13PIQLYY17 pKa = 10.12GALLEE22 pKa = 4.19ICEE25 pKa = 4.12IFAYY29 pKa = 8.25QHH31 pKa = 5.75EE32 pKa = 4.68VAPTTGYY39 pKa = 10.22RR40 pKa = 11.84HH41 pKa = 4.6YY42 pKa = 10.61QGYY45 pKa = 10.34LEE47 pKa = 5.49LVTKK51 pKa = 10.49NRR53 pKa = 11.84HH54 pKa = 3.83TWIQNQLLKK63 pKa = 10.78HH64 pKa = 5.73GLHH67 pKa = 6.47FEE69 pKa = 4.28YY70 pKa = 10.79LVGAKK75 pKa = 9.76GLPKK79 pKa = 10.05QAWAYY84 pKa = 7.26ATKK87 pKa = 10.36EE88 pKa = 4.1DD89 pKa = 4.14TRR91 pKa = 11.84ADD93 pKa = 4.04GPWTFGEE100 pKa = 4.26PRR102 pKa = 11.84DD103 pKa = 3.81SEE105 pKa = 4.36KK106 pKa = 11.27ANKK109 pKa = 9.53SWLFVKK115 pKa = 10.38AIKK118 pKa = 10.52SGATDD123 pKa = 4.67AEE125 pKa = 4.54LCDD128 pKa = 4.31SFPGMMTNSFSNANRR143 pKa = 11.84IRR145 pKa = 11.84QAYY148 pKa = 8.11GVQLPEE154 pKa = 4.19PEE156 pKa = 3.83RR157 pKa = 11.84TVPLEE162 pKa = 3.96LYY164 pKa = 10.48LFYY167 pKa = 11.02GPSGTGKK174 pKa = 9.76SQFARR179 pKa = 11.84DD180 pKa = 3.51QARR183 pKa = 11.84LAKK186 pKa = 9.17MNPYY190 pKa = 9.71VLPIGKK196 pKa = 9.33DD197 pKa = 3.47FWVTPAMCGKK207 pKa = 9.94KK208 pKa = 10.47YY209 pKa = 10.95YY210 pKa = 10.19IIDD213 pKa = 3.98DD214 pKa = 4.26FKK216 pKa = 11.85SNLGLSDD223 pKa = 4.69LLRR226 pKa = 11.84LLDD229 pKa = 4.15TYY231 pKa = 10.9PVEE234 pKa = 4.6APNKK238 pKa = 9.42GIASSEE244 pKa = 3.89FSSLNGGVFNNFVRR258 pKa = 11.84WPYY261 pKa = 10.87VVDD264 pKa = 3.21ARR266 pKa = 11.84NYY268 pKa = 8.35CHH270 pKa = 6.75YY271 pKa = 9.77NQSITSS277 pKa = 3.6
Molecular weight: 31.3 kDa
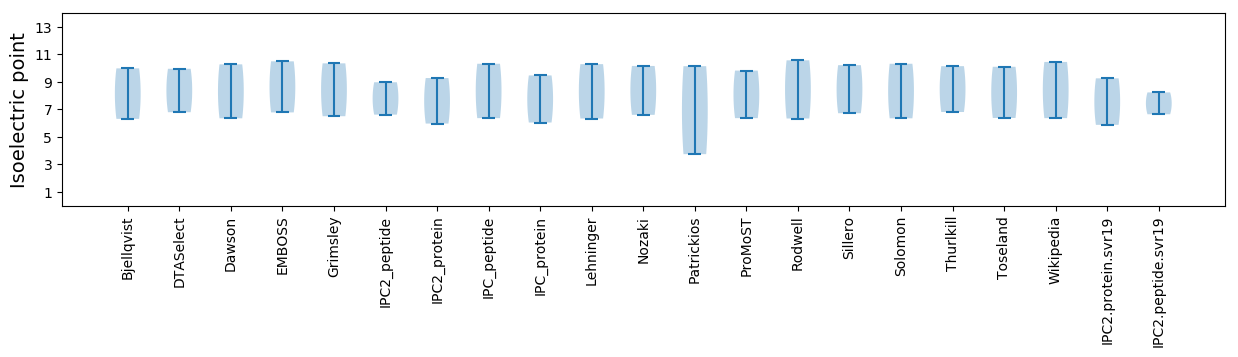
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA95|A0A126GA95_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-29 OX=1685756 PE=4 SV=1
MM1 pKa = 7.05CTHH4 pKa = 6.87ARR6 pKa = 11.84TWMPCRR12 pKa = 11.84KK13 pKa = 7.65MARR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84VTRR21 pKa = 11.84KK22 pKa = 8.22MKK24 pKa = 10.4KK25 pKa = 8.54PRR27 pKa = 11.84ASARR31 pKa = 11.84RR32 pKa = 11.84NPTQEE37 pKa = 4.59FLMKK41 pKa = 9.83PAIRR45 pKa = 11.84ARR47 pKa = 11.84GSRR50 pKa = 11.84SKK52 pKa = 10.69RR53 pKa = 11.84QGYY56 pKa = 8.34MSAAMGTGLMPCTKK70 pKa = 9.76RR71 pKa = 11.84YY72 pKa = 8.41MDD74 pKa = 5.58SILDD78 pKa = 3.7PSGDD82 pKa = 3.69AGRR85 pKa = 11.84GACVPSGFPIPSQKK99 pKa = 10.31ARR101 pKa = 11.84AFLRR105 pKa = 11.84GTMSTGTGGNGYY117 pKa = 9.57ILFTPSLGNDD127 pKa = 3.05NAIVRR132 pKa = 11.84FTTAANTAVGTEE144 pKa = 3.84ALNNAVYY151 pKa = 8.6TASNSSISAGKK162 pKa = 10.09LPYY165 pKa = 9.32STAQIASGNIDD176 pKa = 3.32ARR178 pKa = 11.84IVSAMIRR185 pKa = 11.84VRR187 pKa = 11.84FAGAEE192 pKa = 4.07DD193 pKa = 3.71NRR195 pKa = 11.84SGIVTLLEE203 pKa = 4.86DD204 pKa = 4.88PDD206 pKa = 4.35HH207 pKa = 7.0NSLFTQTPAQVASFEE222 pKa = 4.26GATRR226 pKa = 11.84EE227 pKa = 4.06RR228 pKa = 11.84PNGDD232 pKa = 3.5GAWSQICWSGPCKK245 pKa = 8.83QTEE248 pKa = 4.11VEE250 pKa = 4.43YY251 pKa = 11.42VNTATQTTQTDD262 pKa = 2.74IMAILINGTTNSAGAPGPQPFEE284 pKa = 3.76YY285 pKa = 10.01EE286 pKa = 3.38AWINCEE292 pKa = 3.94YY293 pKa = 10.34IGRR296 pKa = 11.84DD297 pKa = 3.78AIGKK301 pKa = 8.04TDD303 pKa = 3.54NQNDD307 pKa = 3.87QMGMDD312 pKa = 3.8AVTSIFKK319 pKa = 10.53KK320 pKa = 10.41FAGTPQVQVNPEE332 pKa = 4.03SAPSIRR338 pKa = 11.84QAIRR342 pKa = 11.84THH344 pKa = 5.56QMSVSGSALPRR355 pKa = 11.84PKK357 pKa = 9.86TLAGDD362 pKa = 4.67LIEE365 pKa = 4.66GAVTAFNPGWGRR377 pKa = 11.84LWGSLRR383 pKa = 11.84DD384 pKa = 3.76RR385 pKa = 11.84IRR387 pKa = 11.84ARR389 pKa = 11.84KK390 pKa = 8.65
MM1 pKa = 7.05CTHH4 pKa = 6.87ARR6 pKa = 11.84TWMPCRR12 pKa = 11.84KK13 pKa = 7.65MARR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84VTRR21 pKa = 11.84KK22 pKa = 8.22MKK24 pKa = 10.4KK25 pKa = 8.54PRR27 pKa = 11.84ASARR31 pKa = 11.84RR32 pKa = 11.84NPTQEE37 pKa = 4.59FLMKK41 pKa = 9.83PAIRR45 pKa = 11.84ARR47 pKa = 11.84GSRR50 pKa = 11.84SKK52 pKa = 10.69RR53 pKa = 11.84QGYY56 pKa = 8.34MSAAMGTGLMPCTKK70 pKa = 9.76RR71 pKa = 11.84YY72 pKa = 8.41MDD74 pKa = 5.58SILDD78 pKa = 3.7PSGDD82 pKa = 3.69AGRR85 pKa = 11.84GACVPSGFPIPSQKK99 pKa = 10.31ARR101 pKa = 11.84AFLRR105 pKa = 11.84GTMSTGTGGNGYY117 pKa = 9.57ILFTPSLGNDD127 pKa = 3.05NAIVRR132 pKa = 11.84FTTAANTAVGTEE144 pKa = 3.84ALNNAVYY151 pKa = 8.6TASNSSISAGKK162 pKa = 10.09LPYY165 pKa = 9.32STAQIASGNIDD176 pKa = 3.32ARR178 pKa = 11.84IVSAMIRR185 pKa = 11.84VRR187 pKa = 11.84FAGAEE192 pKa = 4.07DD193 pKa = 3.71NRR195 pKa = 11.84SGIVTLLEE203 pKa = 4.86DD204 pKa = 4.88PDD206 pKa = 4.35HH207 pKa = 7.0NSLFTQTPAQVASFEE222 pKa = 4.26GATRR226 pKa = 11.84EE227 pKa = 4.06RR228 pKa = 11.84PNGDD232 pKa = 3.5GAWSQICWSGPCKK245 pKa = 8.83QTEE248 pKa = 4.11VEE250 pKa = 4.43YY251 pKa = 11.42VNTATQTTQTDD262 pKa = 2.74IMAILINGTTNSAGAPGPQPFEE284 pKa = 3.76YY285 pKa = 10.01EE286 pKa = 3.38AWINCEE292 pKa = 3.94YY293 pKa = 10.34IGRR296 pKa = 11.84DD297 pKa = 3.78AIGKK301 pKa = 8.04TDD303 pKa = 3.54NQNDD307 pKa = 3.87QMGMDD312 pKa = 3.8AVTSIFKK319 pKa = 10.53KK320 pKa = 10.41FAGTPQVQVNPEE332 pKa = 4.03SAPSIRR338 pKa = 11.84QAIRR342 pKa = 11.84THH344 pKa = 5.56QMSVSGSALPRR355 pKa = 11.84PKK357 pKa = 9.86TLAGDD362 pKa = 4.67LIEE365 pKa = 4.66GAVTAFNPGWGRR377 pKa = 11.84LWGSLRR383 pKa = 11.84DD384 pKa = 3.76RR385 pKa = 11.84IRR387 pKa = 11.84ARR389 pKa = 11.84KK390 pKa = 8.65
Molecular weight: 42.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
667 |
277 |
390 |
333.5 |
36.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.195 ± 1.19 | 1.649 ± 0.129 |
4.498 ± 0.35 | 4.348 ± 0.898 |
3.898 ± 0.727 | 8.546 ± 0.607 |
1.349 ± 0.514 | 5.097 ± 0.708 |
4.648 ± 0.71 | 6.297 ± 1.716 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.999 ± 0.751 | 5.097 ± 0.027 |
5.997 ± 0.139 | 4.198 ± 0.143 |
6.747 ± 1.519 | 7.196 ± 0.439 |
7.196 ± 1.347 | 4.498 ± 0.35 |
1.799 ± 0.231 | 3.748 ± 1.503 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
