
Maize-associated totivirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
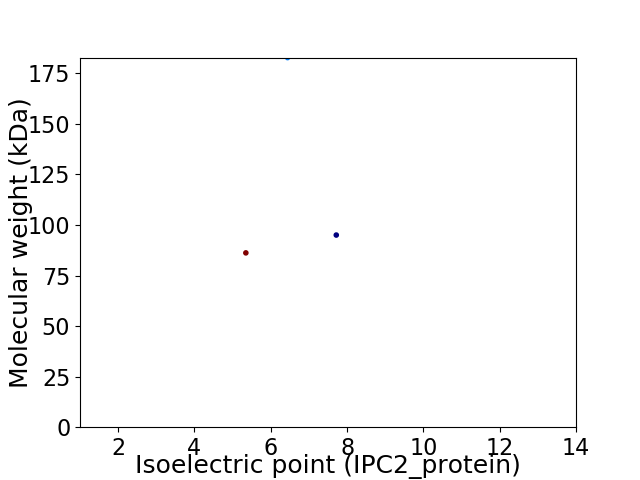
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
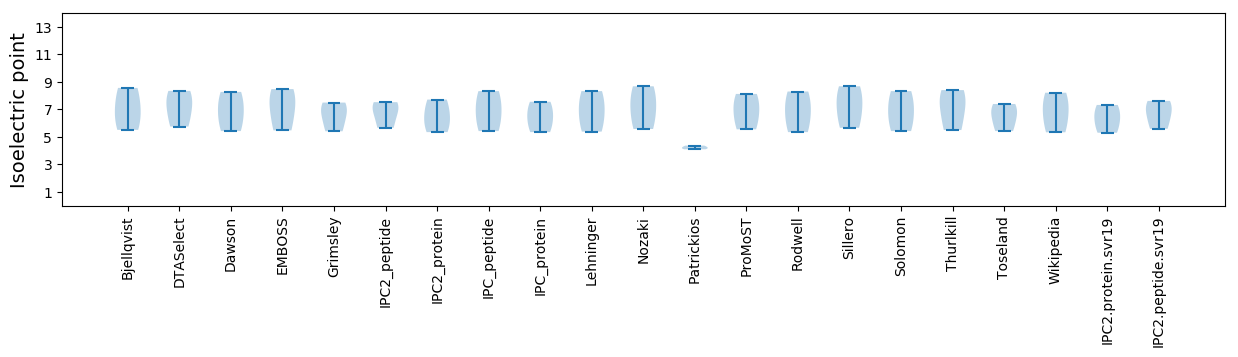
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A193GXV6|A0A193GXV6_9VIRU Coat protein OS=Maize-associated totivirus 2 OX=1874267 PE=4 SV=2
MM1 pKa = 7.59DD2 pKa = 3.86TFVRR6 pKa = 11.84HH7 pKa = 5.63FFKK10 pKa = 10.78EE11 pKa = 4.03VTTPGVPFVLDD22 pKa = 4.21DD23 pKa = 4.33ACHH26 pKa = 5.44TWTYY30 pKa = 10.17FQGFKK35 pKa = 10.27TVTTGDD41 pKa = 3.41ATKK44 pKa = 10.77SSARR48 pKa = 11.84DD49 pKa = 3.17IYY51 pKa = 11.26QAMTPLGMLDD61 pKa = 3.34AAFEE65 pKa = 4.8SMGSNYY71 pKa = 10.7DD72 pKa = 3.6GLTKK76 pKa = 10.65NYY78 pKa = 7.33LTPEE82 pKa = 3.95GAINFSNIHH91 pKa = 6.26DD92 pKa = 4.01TLKK95 pKa = 10.89QSGGMNNQDD104 pKa = 3.96LSTFMAFCQTISWHH118 pKa = 6.73DD119 pKa = 3.44NHH121 pKa = 5.78VTLLFNMLRR130 pKa = 11.84YY131 pKa = 9.76VILMKK136 pKa = 10.81FKK138 pKa = 10.51EE139 pKa = 4.58CKK141 pKa = 9.74QNLNGALGRR150 pKa = 11.84FQEE153 pKa = 5.08DD154 pKa = 3.44FLEE157 pKa = 5.15INMDD161 pKa = 3.75QYY163 pKa = 11.43WPDD166 pKa = 3.29SYY168 pKa = 11.53PRR170 pKa = 11.84DD171 pKa = 3.73CAFDD175 pKa = 3.44SWPVDD180 pKa = 3.5VTEE183 pKa = 5.48DD184 pKa = 3.26GAIALEE190 pKa = 4.53HH191 pKa = 6.41LTTDD195 pKa = 3.15QTRR198 pKa = 11.84NIGPAIDD205 pKa = 4.35LRR207 pKa = 11.84GLTSAEE213 pKa = 4.19SIFVIFMLHH222 pKa = 4.92KK223 pKa = 7.64WTRR226 pKa = 11.84SSRR229 pKa = 11.84FKK231 pKa = 11.11LDD233 pKa = 2.99VSTKK237 pKa = 10.71LLTNTLQFRR246 pKa = 11.84GVDD249 pKa = 3.35VSALYY254 pKa = 10.42DD255 pKa = 3.9SYY257 pKa = 11.06WKK259 pKa = 10.73EE260 pKa = 3.97EE261 pKa = 4.11TVMPALLSSNAAWHH275 pKa = 6.52ALAKK279 pKa = 10.59YY280 pKa = 7.53VTQNRR285 pKa = 11.84LFDD288 pKa = 3.71QFSSALYY295 pKa = 10.36LVTALTYY302 pKa = 10.44QYY304 pKa = 11.16VPEE307 pKa = 4.19TAEE310 pKa = 4.24GVSWLSLNWRR320 pKa = 11.84VVLPEE325 pKa = 3.81FTSIRR330 pKa = 11.84GKK332 pKa = 10.35FAFFNTGDD340 pKa = 3.66AAFLSQRR347 pKa = 11.84SLDD350 pKa = 3.2EE351 pKa = 3.74WEE353 pKa = 4.37YY354 pKa = 11.01IGGKK358 pKa = 7.04MEE360 pKa = 5.39KK361 pKa = 9.65INLMSLTFCQAFYY374 pKa = 11.16AGIALRR380 pKa = 11.84TTRR383 pKa = 11.84RR384 pKa = 11.84GLEE387 pKa = 3.85MMPADD392 pKa = 4.17IYY394 pKa = 11.06GTDD397 pKa = 3.36AEE399 pKa = 4.62FWNAEE404 pKa = 4.15NFVSAAVAEE413 pKa = 4.58FARR416 pKa = 11.84IKK418 pKa = 10.79VPMSGTPGLYY428 pKa = 8.37ITCDD432 pKa = 3.2EE433 pKa = 4.64RR434 pKa = 11.84LDD436 pKa = 3.58IVNEE440 pKa = 3.79HH441 pKa = 6.45RR442 pKa = 11.84FVMTNNPNRR451 pKa = 11.84RR452 pKa = 11.84DD453 pKa = 3.65LPPTHH458 pKa = 7.23TYY460 pKa = 11.41GEE462 pKa = 4.48TQTNEE467 pKa = 4.06TVVDD471 pKa = 5.03DD472 pKa = 4.87IEE474 pKa = 5.74LDD476 pKa = 3.65PTTLPDD482 pKa = 4.64RR483 pKa = 11.84DD484 pKa = 4.08PEE486 pKa = 4.13KK487 pKa = 10.49QALINGMRR495 pKa = 11.84VAKK498 pKa = 10.46VGAEE502 pKa = 3.92AMRR505 pKa = 11.84AVDD508 pKa = 4.51PEE510 pKa = 4.27CSLPAGFEE518 pKa = 3.62LALRR522 pKa = 11.84DD523 pKa = 3.25RR524 pKa = 11.84VAARR528 pKa = 11.84RR529 pKa = 11.84PRR531 pKa = 11.84VRR533 pKa = 11.84QAARR537 pKa = 11.84VTVACPLLPFPGVPTLLLPTQCTPYY562 pKa = 10.35PIPFEE567 pKa = 4.04LAGKK571 pKa = 10.23FDD573 pKa = 5.08DD574 pKa = 3.74NLGRR578 pKa = 11.84FDD580 pKa = 4.74RR581 pKa = 11.84RR582 pKa = 11.84GTRR585 pKa = 11.84LKK587 pKa = 10.13IDD589 pKa = 3.58AAWRR593 pKa = 11.84VMNFLQDD600 pKa = 3.54QSYY603 pKa = 8.1TWEE606 pKa = 3.98CEE608 pKa = 3.82YY609 pKa = 10.85RR610 pKa = 11.84GTRR613 pKa = 11.84IGQQEE618 pKa = 4.44YY619 pKa = 10.05VRR621 pKa = 11.84GRR623 pKa = 11.84YY624 pKa = 9.93SNMIWPVLYY633 pKa = 10.44QPEE636 pKa = 4.66DD637 pKa = 3.51NDD639 pKa = 3.26EE640 pKa = 4.28DD641 pKa = 4.11VVLRR645 pKa = 11.84RR646 pKa = 11.84ITEE649 pKa = 3.81VGLPDD654 pKa = 3.66VPLPPMHH661 pKa = 6.93NEE663 pKa = 3.74FFKK666 pKa = 11.02GRR668 pKa = 11.84SIEE671 pKa = 3.95YY672 pKa = 9.04KK673 pKa = 8.21YY674 pKa = 10.98QSIRR678 pKa = 11.84RR679 pKa = 11.84GLATRR684 pKa = 11.84FRR686 pKa = 11.84GAKK689 pKa = 9.23IDD691 pKa = 3.37ISSYY695 pKa = 8.68GAHH698 pKa = 5.7VPVLKK703 pKa = 10.57KK704 pKa = 10.46ISTVTYY710 pKa = 10.22AVNEE714 pKa = 4.09GLRR717 pKa = 11.84TVRR720 pKa = 11.84GFIEE724 pKa = 4.44RR725 pKa = 11.84AAQDD729 pKa = 3.36FRR731 pKa = 11.84LAPVAGPGVIPPVEE745 pKa = 3.96QAPDD749 pKa = 3.14ASAATGGQEE758 pKa = 4.33TGGTSAA764 pKa = 5.04
MM1 pKa = 7.59DD2 pKa = 3.86TFVRR6 pKa = 11.84HH7 pKa = 5.63FFKK10 pKa = 10.78EE11 pKa = 4.03VTTPGVPFVLDD22 pKa = 4.21DD23 pKa = 4.33ACHH26 pKa = 5.44TWTYY30 pKa = 10.17FQGFKK35 pKa = 10.27TVTTGDD41 pKa = 3.41ATKK44 pKa = 10.77SSARR48 pKa = 11.84DD49 pKa = 3.17IYY51 pKa = 11.26QAMTPLGMLDD61 pKa = 3.34AAFEE65 pKa = 4.8SMGSNYY71 pKa = 10.7DD72 pKa = 3.6GLTKK76 pKa = 10.65NYY78 pKa = 7.33LTPEE82 pKa = 3.95GAINFSNIHH91 pKa = 6.26DD92 pKa = 4.01TLKK95 pKa = 10.89QSGGMNNQDD104 pKa = 3.96LSTFMAFCQTISWHH118 pKa = 6.73DD119 pKa = 3.44NHH121 pKa = 5.78VTLLFNMLRR130 pKa = 11.84YY131 pKa = 9.76VILMKK136 pKa = 10.81FKK138 pKa = 10.51EE139 pKa = 4.58CKK141 pKa = 9.74QNLNGALGRR150 pKa = 11.84FQEE153 pKa = 5.08DD154 pKa = 3.44FLEE157 pKa = 5.15INMDD161 pKa = 3.75QYY163 pKa = 11.43WPDD166 pKa = 3.29SYY168 pKa = 11.53PRR170 pKa = 11.84DD171 pKa = 3.73CAFDD175 pKa = 3.44SWPVDD180 pKa = 3.5VTEE183 pKa = 5.48DD184 pKa = 3.26GAIALEE190 pKa = 4.53HH191 pKa = 6.41LTTDD195 pKa = 3.15QTRR198 pKa = 11.84NIGPAIDD205 pKa = 4.35LRR207 pKa = 11.84GLTSAEE213 pKa = 4.19SIFVIFMLHH222 pKa = 4.92KK223 pKa = 7.64WTRR226 pKa = 11.84SSRR229 pKa = 11.84FKK231 pKa = 11.11LDD233 pKa = 2.99VSTKK237 pKa = 10.71LLTNTLQFRR246 pKa = 11.84GVDD249 pKa = 3.35VSALYY254 pKa = 10.42DD255 pKa = 3.9SYY257 pKa = 11.06WKK259 pKa = 10.73EE260 pKa = 3.97EE261 pKa = 4.11TVMPALLSSNAAWHH275 pKa = 6.52ALAKK279 pKa = 10.59YY280 pKa = 7.53VTQNRR285 pKa = 11.84LFDD288 pKa = 3.71QFSSALYY295 pKa = 10.36LVTALTYY302 pKa = 10.44QYY304 pKa = 11.16VPEE307 pKa = 4.19TAEE310 pKa = 4.24GVSWLSLNWRR320 pKa = 11.84VVLPEE325 pKa = 3.81FTSIRR330 pKa = 11.84GKK332 pKa = 10.35FAFFNTGDD340 pKa = 3.66AAFLSQRR347 pKa = 11.84SLDD350 pKa = 3.2EE351 pKa = 3.74WEE353 pKa = 4.37YY354 pKa = 11.01IGGKK358 pKa = 7.04MEE360 pKa = 5.39KK361 pKa = 9.65INLMSLTFCQAFYY374 pKa = 11.16AGIALRR380 pKa = 11.84TTRR383 pKa = 11.84RR384 pKa = 11.84GLEE387 pKa = 3.85MMPADD392 pKa = 4.17IYY394 pKa = 11.06GTDD397 pKa = 3.36AEE399 pKa = 4.62FWNAEE404 pKa = 4.15NFVSAAVAEE413 pKa = 4.58FARR416 pKa = 11.84IKK418 pKa = 10.79VPMSGTPGLYY428 pKa = 8.37ITCDD432 pKa = 3.2EE433 pKa = 4.64RR434 pKa = 11.84LDD436 pKa = 3.58IVNEE440 pKa = 3.79HH441 pKa = 6.45RR442 pKa = 11.84FVMTNNPNRR451 pKa = 11.84RR452 pKa = 11.84DD453 pKa = 3.65LPPTHH458 pKa = 7.23TYY460 pKa = 11.41GEE462 pKa = 4.48TQTNEE467 pKa = 4.06TVVDD471 pKa = 5.03DD472 pKa = 4.87IEE474 pKa = 5.74LDD476 pKa = 3.65PTTLPDD482 pKa = 4.64RR483 pKa = 11.84DD484 pKa = 4.08PEE486 pKa = 4.13KK487 pKa = 10.49QALINGMRR495 pKa = 11.84VAKK498 pKa = 10.46VGAEE502 pKa = 3.92AMRR505 pKa = 11.84AVDD508 pKa = 4.51PEE510 pKa = 4.27CSLPAGFEE518 pKa = 3.62LALRR522 pKa = 11.84DD523 pKa = 3.25RR524 pKa = 11.84VAARR528 pKa = 11.84RR529 pKa = 11.84PRR531 pKa = 11.84VRR533 pKa = 11.84QAARR537 pKa = 11.84VTVACPLLPFPGVPTLLLPTQCTPYY562 pKa = 10.35PIPFEE567 pKa = 4.04LAGKK571 pKa = 10.23FDD573 pKa = 5.08DD574 pKa = 3.74NLGRR578 pKa = 11.84FDD580 pKa = 4.74RR581 pKa = 11.84RR582 pKa = 11.84GTRR585 pKa = 11.84LKK587 pKa = 10.13IDD589 pKa = 3.58AAWRR593 pKa = 11.84VMNFLQDD600 pKa = 3.54QSYY603 pKa = 8.1TWEE606 pKa = 3.98CEE608 pKa = 3.82YY609 pKa = 10.85RR610 pKa = 11.84GTRR613 pKa = 11.84IGQQEE618 pKa = 4.44YY619 pKa = 10.05VRR621 pKa = 11.84GRR623 pKa = 11.84YY624 pKa = 9.93SNMIWPVLYY633 pKa = 10.44QPEE636 pKa = 4.66DD637 pKa = 3.51NDD639 pKa = 3.26EE640 pKa = 4.28DD641 pKa = 4.11VVLRR645 pKa = 11.84RR646 pKa = 11.84ITEE649 pKa = 3.81VGLPDD654 pKa = 3.66VPLPPMHH661 pKa = 6.93NEE663 pKa = 3.74FFKK666 pKa = 11.02GRR668 pKa = 11.84SIEE671 pKa = 3.95YY672 pKa = 9.04KK673 pKa = 8.21YY674 pKa = 10.98QSIRR678 pKa = 11.84RR679 pKa = 11.84GLATRR684 pKa = 11.84FRR686 pKa = 11.84GAKK689 pKa = 9.23IDD691 pKa = 3.37ISSYY695 pKa = 8.68GAHH698 pKa = 5.7VPVLKK703 pKa = 10.57KK704 pKa = 10.46ISTVTYY710 pKa = 10.22AVNEE714 pKa = 4.09GLRR717 pKa = 11.84TVRR720 pKa = 11.84GFIEE724 pKa = 4.44RR725 pKa = 11.84AAQDD729 pKa = 3.36FRR731 pKa = 11.84LAPVAGPGVIPPVEE745 pKa = 3.96QAPDD749 pKa = 3.14ASAATGGQEE758 pKa = 4.33TGGTSAA764 pKa = 5.04
Molecular weight: 86.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A193GXK6|A0A193GXK6_9VIRU RNA-directed RNA polymerase OS=Maize-associated totivirus 2 OX=1874267 PE=3 SV=2
MM1 pKa = 7.59LNATPGGDD9 pKa = 3.23GPYY12 pKa = 9.95IKK14 pKa = 10.19RR15 pKa = 11.84KK16 pKa = 9.8EE17 pKa = 4.04PLTLNNIPVWVNIRR31 pKa = 11.84HH32 pKa = 5.18NHH34 pKa = 4.33LTAVDD39 pKa = 3.51RR40 pKa = 11.84HH41 pKa = 5.28DD42 pKa = 3.56AKK44 pKa = 11.12YY45 pKa = 10.76VLGFILPGQSFEE57 pKa = 4.06VGGFGRR63 pKa = 11.84YY64 pKa = 8.25LVGKK68 pKa = 7.45TCVAAVAIPCDD79 pKa = 4.17GMWLLYY85 pKa = 10.92VSVTTPANILPPIVKK100 pKa = 8.62RR101 pKa = 11.84TLSAMFSSVDD111 pKa = 3.01GYY113 pKa = 11.28YY114 pKa = 10.48FHH116 pKa = 8.03DD117 pKa = 4.89ADD119 pKa = 3.74TTKK122 pKa = 10.91YY123 pKa = 10.19LRR125 pKa = 11.84KK126 pKa = 7.98TFDD129 pKa = 3.23VEE131 pKa = 4.18RR132 pKa = 11.84SAIAHH137 pKa = 6.09AAKK140 pKa = 9.96PIPPRR145 pKa = 11.84EE146 pKa = 4.28GEE148 pKa = 3.98FEE150 pKa = 4.03RR151 pKa = 11.84AVITAEE157 pKa = 3.76HH158 pKa = 6.0HH159 pKa = 3.99THH161 pKa = 5.67YY162 pKa = 10.47RR163 pKa = 11.84PEE165 pKa = 4.48EE166 pKa = 3.43IWAIAEE172 pKa = 3.99QHH174 pKa = 4.93TTTLRR179 pKa = 11.84AMGLVLANLRR189 pKa = 11.84KK190 pKa = 9.53IQGITEE196 pKa = 4.02ATVATFLAYY205 pKa = 9.84IMTVRR210 pKa = 11.84PQVAYY215 pKa = 10.38LVATSKK221 pKa = 10.96RR222 pKa = 11.84IWRR225 pKa = 11.84SKK227 pKa = 10.9NITEE231 pKa = 3.89LTEE234 pKa = 3.83ILKK237 pKa = 10.7EE238 pKa = 3.4IATPIKK244 pKa = 10.62SMHH247 pKa = 4.83QHH249 pKa = 6.84EE250 pKa = 4.73ICDD253 pKa = 3.69LTEE256 pKa = 4.52LFEE259 pKa = 4.38LQCLVNRR266 pKa = 11.84GVGQIDD272 pKa = 3.16WRR274 pKa = 11.84KK275 pKa = 8.69EE276 pKa = 3.49RR277 pKa = 11.84SHH279 pKa = 6.05RR280 pKa = 11.84TRR282 pKa = 11.84PDD284 pKa = 3.12VVKK287 pKa = 10.92VSIEE291 pKa = 4.13DD292 pKa = 3.48VVKK295 pKa = 10.3YY296 pKa = 9.28ATEE299 pKa = 3.74IFLLGKK305 pKa = 9.33SHH307 pKa = 6.83GYY309 pKa = 10.02HH310 pKa = 5.13YY311 pKa = 11.21RR312 pKa = 11.84KK313 pKa = 8.94MDD315 pKa = 3.23KK316 pKa = 10.2QKK318 pKa = 10.82YY319 pKa = 9.23ISARR323 pKa = 11.84WEE325 pKa = 3.68WSPTGSIHH333 pKa = 5.86SQYY336 pKa = 10.83PEE338 pKa = 4.33DD339 pKa = 3.95EE340 pKa = 4.35PYY342 pKa = 10.21IPKK345 pKa = 10.43NYY347 pKa = 8.79RR348 pKa = 11.84QKK350 pKa = 10.03TKK352 pKa = 10.42FVALNMMSRR361 pKa = 11.84QHH363 pKa = 6.35VDD365 pKa = 2.47SMFLRR370 pKa = 11.84KK371 pKa = 9.52PEE373 pKa = 3.62IHH375 pKa = 6.53AWTSVKK381 pKa = 10.73YY382 pKa = 9.62EE383 pKa = 3.87WAKK386 pKa = 10.25QRR388 pKa = 11.84AIYY391 pKa = 10.09GVDD394 pKa = 3.3LTSTVITNFAMYY406 pKa = 10.0RR407 pKa = 11.84CEE409 pKa = 4.02EE410 pKa = 4.23VFKK413 pKa = 11.11HH414 pKa = 5.86RR415 pKa = 11.84FPVGEE420 pKa = 3.83EE421 pKa = 3.42AAADD425 pKa = 3.84RR426 pKa = 11.84VHH428 pKa = 6.86RR429 pKa = 11.84RR430 pKa = 11.84LKK432 pKa = 10.67IMLEE436 pKa = 4.24DD437 pKa = 4.03NEE439 pKa = 4.66SFCYY443 pKa = 10.81DD444 pKa = 3.25FDD446 pKa = 5.14DD447 pKa = 5.86FNAQHH452 pKa = 6.09STQAMQAVLVAYY464 pKa = 9.0FNVFQADD471 pKa = 3.29MSDD474 pKa = 3.82DD475 pKa = 3.51QRR477 pKa = 11.84EE478 pKa = 3.98AMLWVIDD485 pKa = 3.96SLSDD489 pKa = 3.6VTIHH493 pKa = 6.46NSNVQPPEE501 pKa = 3.75QYY503 pKa = 9.89EE504 pKa = 4.25LKK506 pKa = 9.94GTLLSGWRR514 pKa = 11.84LTTFMNTALNYY525 pKa = 10.15VYY527 pKa = 10.1FKK529 pKa = 11.06AAGCFDD535 pKa = 3.23IGGVRR540 pKa = 11.84DD541 pKa = 3.85SVDD544 pKa = 3.1NGDD547 pKa = 4.47DD548 pKa = 3.37VLVSIKK554 pKa = 10.59HH555 pKa = 5.48IGAAVRR561 pKa = 11.84IHH563 pKa = 6.5HH564 pKa = 6.82RR565 pKa = 11.84MALINARR572 pKa = 11.84AQPTKK577 pKa = 10.89CNVFSVGEE585 pKa = 3.91FLRR588 pKa = 11.84VEE590 pKa = 4.23HH591 pKa = 7.01KK592 pKa = 10.46VDD594 pKa = 3.32KK595 pKa = 10.22DD596 pKa = 3.38TGLGAQYY603 pKa = 9.02LTRR606 pKa = 11.84ACATLVHH613 pKa = 6.2SRR615 pKa = 11.84CEE617 pKa = 4.19SQEE620 pKa = 4.0PEE622 pKa = 4.31HH623 pKa = 6.18LTGAVKK629 pKa = 10.65AIVTRR634 pKa = 11.84ARR636 pKa = 11.84EE637 pKa = 3.86VFEE640 pKa = 4.02RR641 pKa = 11.84AKK643 pKa = 10.44ISQALLADD651 pKa = 4.33LVRR654 pKa = 11.84SAIRR658 pKa = 11.84RR659 pKa = 11.84AATIFHH665 pKa = 6.86RR666 pKa = 11.84PAKK669 pKa = 9.24EE670 pKa = 3.54AFIIAEE676 pKa = 4.0LHH678 pKa = 5.81AVVGGASTDD687 pKa = 3.51DD688 pKa = 3.63FAPIDD693 pKa = 3.69FKK695 pKa = 11.3INEE698 pKa = 3.88RR699 pKa = 11.84CEE701 pKa = 3.98YY702 pKa = 10.86DD703 pKa = 4.33KK704 pKa = 11.21EE705 pKa = 4.06RR706 pKa = 11.84CANDD710 pKa = 3.56KK711 pKa = 11.1NDD713 pKa = 3.61MVVTQDD719 pKa = 4.21LYY721 pKa = 10.84PGIFDD726 pKa = 3.83YY727 pKa = 11.32ARR729 pKa = 11.84TLAQAYY735 pKa = 9.23EE736 pKa = 4.37GVLDD740 pKa = 3.81EE741 pKa = 4.31QQAKK745 pKa = 9.91RR746 pKa = 11.84RR747 pKa = 11.84VISATTRR754 pKa = 11.84QISVTRR760 pKa = 11.84KK761 pKa = 6.37TWLDD765 pKa = 3.59IVPLAADD772 pKa = 3.79TFYY775 pKa = 11.05RR776 pKa = 11.84YY777 pKa = 9.68GRR779 pKa = 11.84ALYY782 pKa = 10.54KK783 pKa = 9.63MYY785 pKa = 10.33HH786 pKa = 6.28GLVSMPHH793 pKa = 6.11LEE795 pKa = 3.84KK796 pKa = 10.88ARR798 pKa = 11.84FVGIPPIALLDD809 pKa = 3.66SRR811 pKa = 11.84SRR813 pKa = 11.84NMIRR817 pKa = 11.84NIIVEE822 pKa = 4.16ASDD825 pKa = 3.14VEE827 pKa = 4.31YY828 pKa = 10.99ALRR831 pKa = 11.84VLLL834 pKa = 4.86
MM1 pKa = 7.59LNATPGGDD9 pKa = 3.23GPYY12 pKa = 9.95IKK14 pKa = 10.19RR15 pKa = 11.84KK16 pKa = 9.8EE17 pKa = 4.04PLTLNNIPVWVNIRR31 pKa = 11.84HH32 pKa = 5.18NHH34 pKa = 4.33LTAVDD39 pKa = 3.51RR40 pKa = 11.84HH41 pKa = 5.28DD42 pKa = 3.56AKK44 pKa = 11.12YY45 pKa = 10.76VLGFILPGQSFEE57 pKa = 4.06VGGFGRR63 pKa = 11.84YY64 pKa = 8.25LVGKK68 pKa = 7.45TCVAAVAIPCDD79 pKa = 4.17GMWLLYY85 pKa = 10.92VSVTTPANILPPIVKK100 pKa = 8.62RR101 pKa = 11.84TLSAMFSSVDD111 pKa = 3.01GYY113 pKa = 11.28YY114 pKa = 10.48FHH116 pKa = 8.03DD117 pKa = 4.89ADD119 pKa = 3.74TTKK122 pKa = 10.91YY123 pKa = 10.19LRR125 pKa = 11.84KK126 pKa = 7.98TFDD129 pKa = 3.23VEE131 pKa = 4.18RR132 pKa = 11.84SAIAHH137 pKa = 6.09AAKK140 pKa = 9.96PIPPRR145 pKa = 11.84EE146 pKa = 4.28GEE148 pKa = 3.98FEE150 pKa = 4.03RR151 pKa = 11.84AVITAEE157 pKa = 3.76HH158 pKa = 6.0HH159 pKa = 3.99THH161 pKa = 5.67YY162 pKa = 10.47RR163 pKa = 11.84PEE165 pKa = 4.48EE166 pKa = 3.43IWAIAEE172 pKa = 3.99QHH174 pKa = 4.93TTTLRR179 pKa = 11.84AMGLVLANLRR189 pKa = 11.84KK190 pKa = 9.53IQGITEE196 pKa = 4.02ATVATFLAYY205 pKa = 9.84IMTVRR210 pKa = 11.84PQVAYY215 pKa = 10.38LVATSKK221 pKa = 10.96RR222 pKa = 11.84IWRR225 pKa = 11.84SKK227 pKa = 10.9NITEE231 pKa = 3.89LTEE234 pKa = 3.83ILKK237 pKa = 10.7EE238 pKa = 3.4IATPIKK244 pKa = 10.62SMHH247 pKa = 4.83QHH249 pKa = 6.84EE250 pKa = 4.73ICDD253 pKa = 3.69LTEE256 pKa = 4.52LFEE259 pKa = 4.38LQCLVNRR266 pKa = 11.84GVGQIDD272 pKa = 3.16WRR274 pKa = 11.84KK275 pKa = 8.69EE276 pKa = 3.49RR277 pKa = 11.84SHH279 pKa = 6.05RR280 pKa = 11.84TRR282 pKa = 11.84PDD284 pKa = 3.12VVKK287 pKa = 10.92VSIEE291 pKa = 4.13DD292 pKa = 3.48VVKK295 pKa = 10.3YY296 pKa = 9.28ATEE299 pKa = 3.74IFLLGKK305 pKa = 9.33SHH307 pKa = 6.83GYY309 pKa = 10.02HH310 pKa = 5.13YY311 pKa = 11.21RR312 pKa = 11.84KK313 pKa = 8.94MDD315 pKa = 3.23KK316 pKa = 10.2QKK318 pKa = 10.82YY319 pKa = 9.23ISARR323 pKa = 11.84WEE325 pKa = 3.68WSPTGSIHH333 pKa = 5.86SQYY336 pKa = 10.83PEE338 pKa = 4.33DD339 pKa = 3.95EE340 pKa = 4.35PYY342 pKa = 10.21IPKK345 pKa = 10.43NYY347 pKa = 8.79RR348 pKa = 11.84QKK350 pKa = 10.03TKK352 pKa = 10.42FVALNMMSRR361 pKa = 11.84QHH363 pKa = 6.35VDD365 pKa = 2.47SMFLRR370 pKa = 11.84KK371 pKa = 9.52PEE373 pKa = 3.62IHH375 pKa = 6.53AWTSVKK381 pKa = 10.73YY382 pKa = 9.62EE383 pKa = 3.87WAKK386 pKa = 10.25QRR388 pKa = 11.84AIYY391 pKa = 10.09GVDD394 pKa = 3.3LTSTVITNFAMYY406 pKa = 10.0RR407 pKa = 11.84CEE409 pKa = 4.02EE410 pKa = 4.23VFKK413 pKa = 11.11HH414 pKa = 5.86RR415 pKa = 11.84FPVGEE420 pKa = 3.83EE421 pKa = 3.42AAADD425 pKa = 3.84RR426 pKa = 11.84VHH428 pKa = 6.86RR429 pKa = 11.84RR430 pKa = 11.84LKK432 pKa = 10.67IMLEE436 pKa = 4.24DD437 pKa = 4.03NEE439 pKa = 4.66SFCYY443 pKa = 10.81DD444 pKa = 3.25FDD446 pKa = 5.14DD447 pKa = 5.86FNAQHH452 pKa = 6.09STQAMQAVLVAYY464 pKa = 9.0FNVFQADD471 pKa = 3.29MSDD474 pKa = 3.82DD475 pKa = 3.51QRR477 pKa = 11.84EE478 pKa = 3.98AMLWVIDD485 pKa = 3.96SLSDD489 pKa = 3.6VTIHH493 pKa = 6.46NSNVQPPEE501 pKa = 3.75QYY503 pKa = 9.89EE504 pKa = 4.25LKK506 pKa = 9.94GTLLSGWRR514 pKa = 11.84LTTFMNTALNYY525 pKa = 10.15VYY527 pKa = 10.1FKK529 pKa = 11.06AAGCFDD535 pKa = 3.23IGGVRR540 pKa = 11.84DD541 pKa = 3.85SVDD544 pKa = 3.1NGDD547 pKa = 4.47DD548 pKa = 3.37VLVSIKK554 pKa = 10.59HH555 pKa = 5.48IGAAVRR561 pKa = 11.84IHH563 pKa = 6.5HH564 pKa = 6.82RR565 pKa = 11.84MALINARR572 pKa = 11.84AQPTKK577 pKa = 10.89CNVFSVGEE585 pKa = 3.91FLRR588 pKa = 11.84VEE590 pKa = 4.23HH591 pKa = 7.01KK592 pKa = 10.46VDD594 pKa = 3.32KK595 pKa = 10.22DD596 pKa = 3.38TGLGAQYY603 pKa = 9.02LTRR606 pKa = 11.84ACATLVHH613 pKa = 6.2SRR615 pKa = 11.84CEE617 pKa = 4.19SQEE620 pKa = 4.0PEE622 pKa = 4.31HH623 pKa = 6.18LTGAVKK629 pKa = 10.65AIVTRR634 pKa = 11.84ARR636 pKa = 11.84EE637 pKa = 3.86VFEE640 pKa = 4.02RR641 pKa = 11.84AKK643 pKa = 10.44ISQALLADD651 pKa = 4.33LVRR654 pKa = 11.84SAIRR658 pKa = 11.84RR659 pKa = 11.84AATIFHH665 pKa = 6.86RR666 pKa = 11.84PAKK669 pKa = 9.24EE670 pKa = 3.54AFIIAEE676 pKa = 4.0LHH678 pKa = 5.81AVVGGASTDD687 pKa = 3.51DD688 pKa = 3.63FAPIDD693 pKa = 3.69FKK695 pKa = 11.3INEE698 pKa = 3.88RR699 pKa = 11.84CEE701 pKa = 3.98YY702 pKa = 10.86DD703 pKa = 4.33KK704 pKa = 11.21EE705 pKa = 4.06RR706 pKa = 11.84CANDD710 pKa = 3.56KK711 pKa = 11.1NDD713 pKa = 3.61MVVTQDD719 pKa = 4.21LYY721 pKa = 10.84PGIFDD726 pKa = 3.83YY727 pKa = 11.32ARR729 pKa = 11.84TLAQAYY735 pKa = 9.23EE736 pKa = 4.37GVLDD740 pKa = 3.81EE741 pKa = 4.31QQAKK745 pKa = 9.91RR746 pKa = 11.84RR747 pKa = 11.84VISATTRR754 pKa = 11.84QISVTRR760 pKa = 11.84KK761 pKa = 6.37TWLDD765 pKa = 3.59IVPLAADD772 pKa = 3.79TFYY775 pKa = 11.05RR776 pKa = 11.84YY777 pKa = 9.68GRR779 pKa = 11.84ALYY782 pKa = 10.54KK783 pKa = 9.63MYY785 pKa = 10.33HH786 pKa = 6.28GLVSMPHH793 pKa = 6.11LEE795 pKa = 3.84KK796 pKa = 10.88ARR798 pKa = 11.84FVGIPPIALLDD809 pKa = 3.66SRR811 pKa = 11.84SRR813 pKa = 11.84NMIRR817 pKa = 11.84NIIVEE822 pKa = 4.16ASDD825 pKa = 3.14VEE827 pKa = 4.31YY828 pKa = 10.99ALRR831 pKa = 11.84VLLL834 pKa = 4.86
Molecular weight: 95.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3206 |
764 |
1608 |
1068.7 |
121.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.858 ± 0.27 | 1.528 ± 0.106 |
6.02 ± 0.163 | 5.864 ± 0.102 |
4.741 ± 0.352 | 5.521 ± 0.365 |
2.682 ± 0.451 | 5.49 ± 0.478 |
4.367 ± 0.393 | 8.079 ± 0.228 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.682 ± 0.077 | 3.743 ± 0.201 |
4.616 ± 0.368 | 3.462 ± 0.072 |
7.174 ± 0.13 | 5.115 ± 0.081 |
7.112 ± 0.344 | 7.299 ± 0.34 |
1.716 ± 0.099 | 3.93 ± 0.112 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
