
Streptococcus phage Javan139
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
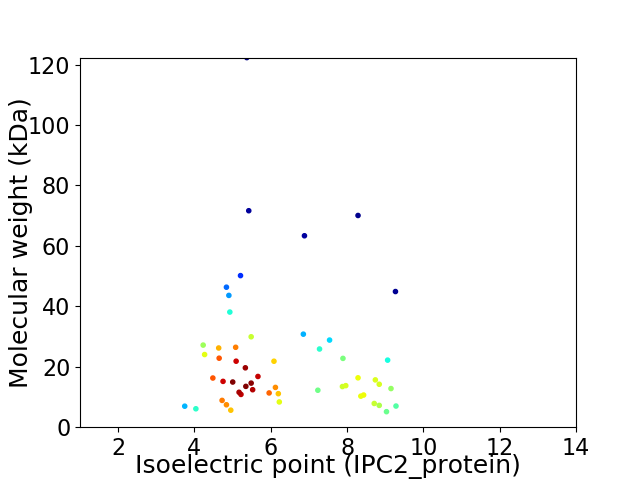
Virtual 2D-PAGE plot for 54 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6A0M4|A0A4D6A0M4_9CAUD Portal protein OS=Streptococcus phage Javan139 OX=2548000 GN=Javan139_0024 PE=4 SV=1
MM1 pKa = 7.49INWNDD6 pKa = 2.94VSLNDD11 pKa = 3.7VICYY15 pKa = 8.94YY16 pKa = 11.36AFVFDD21 pKa = 5.47LDD23 pKa = 3.33ISLVLEE29 pKa = 4.67FSVHH33 pKa = 6.73DD34 pKa = 4.22LDD36 pKa = 6.18EE37 pKa = 4.47IALCKK42 pKa = 10.18LAIEE46 pKa = 4.13NWKK49 pKa = 10.54KK50 pKa = 11.08SEE52 pKa = 3.86
MM1 pKa = 7.49INWNDD6 pKa = 2.94VSLNDD11 pKa = 3.7VICYY15 pKa = 8.94YY16 pKa = 11.36AFVFDD21 pKa = 5.47LDD23 pKa = 3.33ISLVLEE29 pKa = 4.67FSVHH33 pKa = 6.73DD34 pKa = 4.22LDD36 pKa = 6.18EE37 pKa = 4.47IALCKK42 pKa = 10.18LAIEE46 pKa = 4.13NWKK49 pKa = 10.54KK50 pKa = 11.08SEE52 pKa = 3.86
Molecular weight: 6.1 kDa
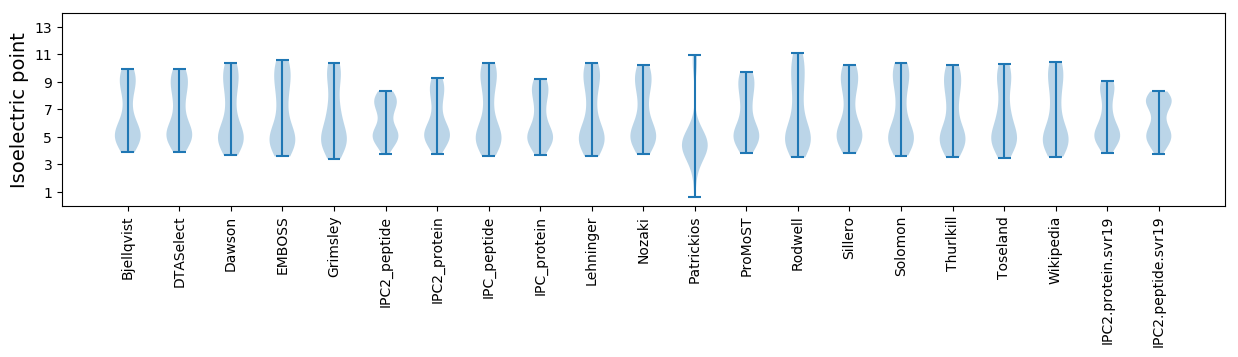
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6A1X8|A0A4D6A1X8_9CAUD Uncharacterized protein OS=Streptococcus phage Javan139 OX=2548000 GN=Javan139_0005 PE=4 SV=1
MM1 pKa = 7.57WIEE4 pKa = 3.88EE5 pKa = 3.99QSNGKK10 pKa = 8.81LRR12 pKa = 11.84YY13 pKa = 7.6VEE15 pKa = 4.67RR16 pKa = 11.84YY17 pKa = 8.82KK18 pKa = 10.81EE19 pKa = 4.19HH20 pKa = 5.75YY21 pKa = 7.4TGKK24 pKa = 10.35YY25 pKa = 7.74KK26 pKa = 10.52RR27 pKa = 11.84AYY29 pKa = 9.59EE30 pKa = 4.42LIEE33 pKa = 4.69RR34 pKa = 11.84DD35 pKa = 3.54TPQGRR40 pKa = 11.84KK41 pKa = 8.92KK42 pKa = 10.56AATALRR48 pKa = 11.84LKK50 pKa = 10.23IEE52 pKa = 4.24KK53 pKa = 9.76LVHH56 pKa = 5.68IRR58 pKa = 11.84PKK60 pKa = 10.25KK61 pKa = 8.81EE62 pKa = 3.4KK63 pKa = 8.23VTFGEE68 pKa = 5.03VYY70 pKa = 10.57QNFYY74 pKa = 11.15SSWSLGVKK82 pKa = 10.01QSTIYY87 pKa = 8.8STKK90 pKa = 10.54NIDD93 pKa = 3.91RR94 pKa = 11.84IILKK98 pKa = 9.95KK99 pKa = 10.38IPNDD103 pKa = 3.46YY104 pKa = 10.59LISKK108 pKa = 9.34IDD110 pKa = 3.32RR111 pKa = 11.84RR112 pKa = 11.84FLQKK116 pKa = 10.37IFDD119 pKa = 4.51DD120 pKa = 4.34LLSEE124 pKa = 4.5GRR126 pKa = 11.84SHH128 pKa = 7.43NYY130 pKa = 8.04VKK132 pKa = 10.48KK133 pKa = 11.1VKK135 pKa = 9.22WKK137 pKa = 9.75LNRR140 pKa = 11.84IFKK143 pKa = 10.15YY144 pKa = 10.08ALRR147 pKa = 11.84MDD149 pKa = 4.56YY150 pKa = 10.79IDD152 pKa = 4.11VNEE155 pKa = 4.64MIHH158 pKa = 6.88VEE160 pKa = 4.12LPKK163 pKa = 10.6EE164 pKa = 3.79ILTAEE169 pKa = 3.93QAKK172 pKa = 10.02KK173 pKa = 10.46KK174 pKa = 10.26KK175 pKa = 10.32EE176 pKa = 3.78KK177 pKa = 10.62FLDD180 pKa = 3.41QKK182 pKa = 10.12EE183 pKa = 4.5FKK185 pKa = 10.84LFAQNLRR192 pKa = 11.84EE193 pKa = 4.1EE194 pKa = 4.51VYY196 pKa = 10.48RR197 pKa = 11.84DD198 pKa = 3.62YY199 pKa = 11.02RR200 pKa = 11.84VKK202 pKa = 10.54KK203 pKa = 8.25YY204 pKa = 10.97LRR206 pKa = 11.84IAIVLYY212 pKa = 8.7LTGMRR217 pKa = 11.84YY218 pKa = 10.29GEE220 pKa = 3.98LAGLNISKK228 pKa = 10.82DD229 pKa = 2.81IDD231 pKa = 3.95FKK233 pKa = 11.44KK234 pKa = 9.62KK235 pKa = 8.9TIHH238 pKa = 6.07IQHH241 pKa = 6.61TFNFRR246 pKa = 11.84TKK248 pKa = 10.47QRR250 pKa = 11.84TTPKK254 pKa = 9.63TKK256 pKa = 10.15GSDD259 pKa = 3.57RR260 pKa = 11.84IIEE263 pKa = 4.14VSDD266 pKa = 3.6TVLKK270 pKa = 10.81NIQEE274 pKa = 4.05QLAEE278 pKa = 4.24NIRR281 pKa = 11.84AGFEE285 pKa = 3.67TDD287 pKa = 3.43YY288 pKa = 11.7LFINTLGYY296 pKa = 6.77PTTPEE301 pKa = 3.68RR302 pKa = 11.84VIGAFKK308 pKa = 10.74RR309 pKa = 11.84HH310 pKa = 4.87GQKK313 pKa = 10.66AGIEE317 pKa = 4.3KK318 pKa = 10.73NITTHH323 pKa = 6.57IFRR326 pKa = 11.84HH327 pKa = 4.67SHH329 pKa = 5.38ISLLAEE335 pKa = 3.99IGIPLPAIMEE345 pKa = 4.41RR346 pKa = 11.84VGHH349 pKa = 6.2SDD351 pKa = 2.74SKK353 pKa = 8.85TTLEE357 pKa = 4.47VYY359 pKa = 10.4SHH361 pKa = 6.15VTLQMTTNLAKK372 pKa = 10.66KK373 pKa = 9.96LNEE376 pKa = 3.98VKK378 pKa = 10.78LFF380 pKa = 3.85
MM1 pKa = 7.57WIEE4 pKa = 3.88EE5 pKa = 3.99QSNGKK10 pKa = 8.81LRR12 pKa = 11.84YY13 pKa = 7.6VEE15 pKa = 4.67RR16 pKa = 11.84YY17 pKa = 8.82KK18 pKa = 10.81EE19 pKa = 4.19HH20 pKa = 5.75YY21 pKa = 7.4TGKK24 pKa = 10.35YY25 pKa = 7.74KK26 pKa = 10.52RR27 pKa = 11.84AYY29 pKa = 9.59EE30 pKa = 4.42LIEE33 pKa = 4.69RR34 pKa = 11.84DD35 pKa = 3.54TPQGRR40 pKa = 11.84KK41 pKa = 8.92KK42 pKa = 10.56AATALRR48 pKa = 11.84LKK50 pKa = 10.23IEE52 pKa = 4.24KK53 pKa = 9.76LVHH56 pKa = 5.68IRR58 pKa = 11.84PKK60 pKa = 10.25KK61 pKa = 8.81EE62 pKa = 3.4KK63 pKa = 8.23VTFGEE68 pKa = 5.03VYY70 pKa = 10.57QNFYY74 pKa = 11.15SSWSLGVKK82 pKa = 10.01QSTIYY87 pKa = 8.8STKK90 pKa = 10.54NIDD93 pKa = 3.91RR94 pKa = 11.84IILKK98 pKa = 9.95KK99 pKa = 10.38IPNDD103 pKa = 3.46YY104 pKa = 10.59LISKK108 pKa = 9.34IDD110 pKa = 3.32RR111 pKa = 11.84RR112 pKa = 11.84FLQKK116 pKa = 10.37IFDD119 pKa = 4.51DD120 pKa = 4.34LLSEE124 pKa = 4.5GRR126 pKa = 11.84SHH128 pKa = 7.43NYY130 pKa = 8.04VKK132 pKa = 10.48KK133 pKa = 11.1VKK135 pKa = 9.22WKK137 pKa = 9.75LNRR140 pKa = 11.84IFKK143 pKa = 10.15YY144 pKa = 10.08ALRR147 pKa = 11.84MDD149 pKa = 4.56YY150 pKa = 10.79IDD152 pKa = 4.11VNEE155 pKa = 4.64MIHH158 pKa = 6.88VEE160 pKa = 4.12LPKK163 pKa = 10.6EE164 pKa = 3.79ILTAEE169 pKa = 3.93QAKK172 pKa = 10.02KK173 pKa = 10.46KK174 pKa = 10.26KK175 pKa = 10.32EE176 pKa = 3.78KK177 pKa = 10.62FLDD180 pKa = 3.41QKK182 pKa = 10.12EE183 pKa = 4.5FKK185 pKa = 10.84LFAQNLRR192 pKa = 11.84EE193 pKa = 4.1EE194 pKa = 4.51VYY196 pKa = 10.48RR197 pKa = 11.84DD198 pKa = 3.62YY199 pKa = 11.02RR200 pKa = 11.84VKK202 pKa = 10.54KK203 pKa = 8.25YY204 pKa = 10.97LRR206 pKa = 11.84IAIVLYY212 pKa = 8.7LTGMRR217 pKa = 11.84YY218 pKa = 10.29GEE220 pKa = 3.98LAGLNISKK228 pKa = 10.82DD229 pKa = 2.81IDD231 pKa = 3.95FKK233 pKa = 11.44KK234 pKa = 9.62KK235 pKa = 8.9TIHH238 pKa = 6.07IQHH241 pKa = 6.61TFNFRR246 pKa = 11.84TKK248 pKa = 10.47QRR250 pKa = 11.84TTPKK254 pKa = 9.63TKK256 pKa = 10.15GSDD259 pKa = 3.57RR260 pKa = 11.84IIEE263 pKa = 4.14VSDD266 pKa = 3.6TVLKK270 pKa = 10.81NIQEE274 pKa = 4.05QLAEE278 pKa = 4.24NIRR281 pKa = 11.84AGFEE285 pKa = 3.67TDD287 pKa = 3.43YY288 pKa = 11.7LFINTLGYY296 pKa = 6.77PTTPEE301 pKa = 3.68RR302 pKa = 11.84VIGAFKK308 pKa = 10.74RR309 pKa = 11.84HH310 pKa = 4.87GQKK313 pKa = 10.66AGIEE317 pKa = 4.3KK318 pKa = 10.73NITTHH323 pKa = 6.57IFRR326 pKa = 11.84HH327 pKa = 4.67SHH329 pKa = 5.38ISLLAEE335 pKa = 3.99IGIPLPAIMEE345 pKa = 4.41RR346 pKa = 11.84VGHH349 pKa = 6.2SDD351 pKa = 2.74SKK353 pKa = 8.85TTLEE357 pKa = 4.47VYY359 pKa = 10.4SHH361 pKa = 6.15VTLQMTTNLAKK372 pKa = 10.66KK373 pKa = 9.96LNEE376 pKa = 3.98VKK378 pKa = 10.78LFF380 pKa = 3.85
Molecular weight: 44.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11044 |
44 |
1133 |
204.5 |
23.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.891 ± 0.455 | 0.561 ± 0.11 |
6.655 ± 0.407 | 7.687 ± 0.314 |
3.903 ± 0.248 | 5.994 ± 0.362 |
1.512 ± 0.156 | 7.905 ± 0.286 |
8.756 ± 0.387 | 8.511 ± 0.225 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.409 ± 0.155 | 5.714 ± 0.263 |
2.608 ± 0.221 | 3.866 ± 0.212 |
4.138 ± 0.297 | 6.112 ± 0.335 |
5.804 ± 0.333 | 5.931 ± 0.28 |
1.177 ± 0.143 | 3.866 ± 0.342 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
