
Thermus virus P23-77
Taxonomy: Viruses; Varidnaviria; Helvetiavirae; Dividoviricota; Laserviricetes; Halopanivirales; Sphaerolipoviridae; Gammasphaerolipovirus
Average proteome isoelectric point is 7.59
Get precalculated fractions of proteins
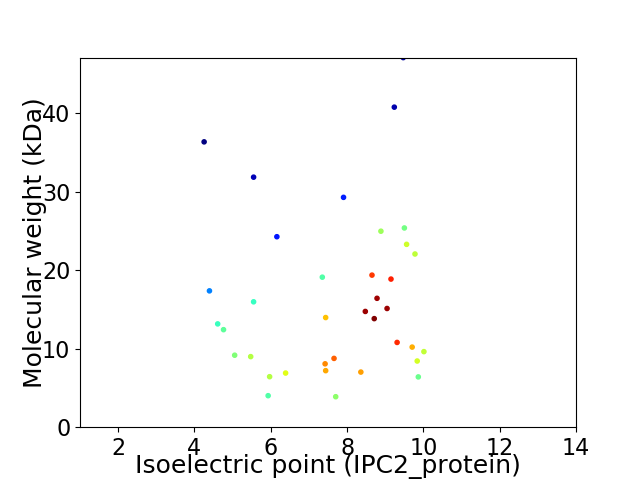
Virtual 2D-PAGE plot for 37 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8CHM7|C8CHM7_9VIRU VP29 OS=Thermus virus P23-77 OX=1714272 PE=4 SV=1
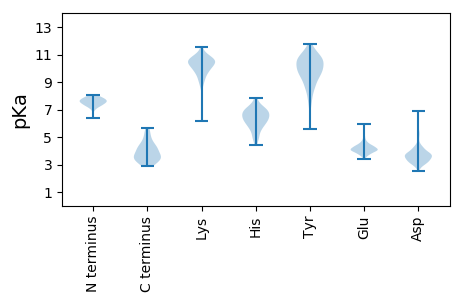
MM1 pKa = 7.96GDD3 pKa = 3.64TGDD6 pKa = 3.69GSYY9 pKa = 9.51LTLYY13 pKa = 10.22GYY15 pKa = 11.21AWLGLLDD22 pKa = 3.74RR23 pKa = 11.84KK24 pKa = 9.4EE25 pKa = 3.76NRR27 pKa = 11.84FRR29 pKa = 11.84LFQAQVPGEE38 pKa = 4.46GPWPLSDD45 pKa = 3.79PRR47 pKa = 11.84GPDD50 pKa = 2.92VAVWVEE56 pKa = 4.13QEE58 pKa = 4.45VPPLPHH64 pKa = 7.11DD65 pKa = 3.99PRR67 pKa = 11.84EE68 pKa = 4.07VRR70 pKa = 11.84HH71 pKa = 5.68MALAFDD77 pKa = 3.51QAARR81 pKa = 11.84HH82 pKa = 4.66VLAYY86 pKa = 10.1EE87 pKa = 3.97RR88 pKa = 11.84GGQVWVRR95 pKa = 11.84QWDD98 pKa = 3.65PVSQVFVMRR107 pKa = 11.84GPFPGVDD114 pKa = 3.1PFLVQDD120 pKa = 3.48ATVGYY125 pKa = 8.43FPPDD129 pKa = 3.19SDD131 pKa = 3.73VLLFHH136 pKa = 7.15LSPDD140 pKa = 3.03RR141 pKa = 11.84TALVMRR147 pKa = 11.84AQRR150 pKa = 11.84EE151 pKa = 4.49LYY153 pKa = 7.85ATPHH157 pKa = 5.82VIEE160 pKa = 4.71TFPSPVVLDD169 pKa = 3.56QAVALPYY176 pKa = 10.29QIEE179 pKa = 4.25LLGSFLDD186 pKa = 3.93ALGEE190 pKa = 4.14TGVVVRR196 pKa = 11.84SEE198 pKa = 4.38VYY200 pKa = 9.22PVRR203 pKa = 11.84VGDD206 pKa = 4.09ALGPATFTAPATGAYY221 pKa = 8.95IPVVVIQDD229 pKa = 4.45LGTDD233 pKa = 3.68TLGTGAFAAPTTGAYY248 pKa = 9.45IPVVVIQDD256 pKa = 4.35LGTEE260 pKa = 4.17ALGSGTFTAPSTGAYY275 pKa = 8.82IPVVVVQDD283 pKa = 4.38LGTEE287 pKa = 4.08ALATGTFTAPATGTYY302 pKa = 9.91VLVVVVQDD310 pKa = 3.92LTASQYY316 pKa = 10.67TDD318 pKa = 2.99GLGYY322 pKa = 10.11EE323 pKa = 4.32ALGTGTFAAPTTGSYY338 pKa = 9.27TLAA341 pKa = 5.26
MM1 pKa = 7.96GDD3 pKa = 3.64TGDD6 pKa = 3.69GSYY9 pKa = 9.51LTLYY13 pKa = 10.22GYY15 pKa = 11.21AWLGLLDD22 pKa = 3.74RR23 pKa = 11.84KK24 pKa = 9.4EE25 pKa = 3.76NRR27 pKa = 11.84FRR29 pKa = 11.84LFQAQVPGEE38 pKa = 4.46GPWPLSDD45 pKa = 3.79PRR47 pKa = 11.84GPDD50 pKa = 2.92VAVWVEE56 pKa = 4.13QEE58 pKa = 4.45VPPLPHH64 pKa = 7.11DD65 pKa = 3.99PRR67 pKa = 11.84EE68 pKa = 4.07VRR70 pKa = 11.84HH71 pKa = 5.68MALAFDD77 pKa = 3.51QAARR81 pKa = 11.84HH82 pKa = 4.66VLAYY86 pKa = 10.1EE87 pKa = 3.97RR88 pKa = 11.84GGQVWVRR95 pKa = 11.84QWDD98 pKa = 3.65PVSQVFVMRR107 pKa = 11.84GPFPGVDD114 pKa = 3.1PFLVQDD120 pKa = 3.48ATVGYY125 pKa = 8.43FPPDD129 pKa = 3.19SDD131 pKa = 3.73VLLFHH136 pKa = 7.15LSPDD140 pKa = 3.03RR141 pKa = 11.84TALVMRR147 pKa = 11.84AQRR150 pKa = 11.84EE151 pKa = 4.49LYY153 pKa = 7.85ATPHH157 pKa = 5.82VIEE160 pKa = 4.71TFPSPVVLDD169 pKa = 3.56QAVALPYY176 pKa = 10.29QIEE179 pKa = 4.25LLGSFLDD186 pKa = 3.93ALGEE190 pKa = 4.14TGVVVRR196 pKa = 11.84SEE198 pKa = 4.38VYY200 pKa = 9.22PVRR203 pKa = 11.84VGDD206 pKa = 4.09ALGPATFTAPATGAYY221 pKa = 8.95IPVVVIQDD229 pKa = 4.45LGTDD233 pKa = 3.68TLGTGAFAAPTTGAYY248 pKa = 9.45IPVVVIQDD256 pKa = 4.35LGTEE260 pKa = 4.17ALGSGTFTAPSTGAYY275 pKa = 8.82IPVVVVQDD283 pKa = 4.38LGTEE287 pKa = 4.08ALATGTFTAPATGTYY302 pKa = 9.91VLVVVVQDD310 pKa = 3.92LTASQYY316 pKa = 10.67TDD318 pKa = 2.99GLGYY322 pKa = 10.11EE323 pKa = 4.32ALGTGTFAAPTTGSYY338 pKa = 9.27TLAA341 pKa = 5.26
Molecular weight: 36.34 kDa
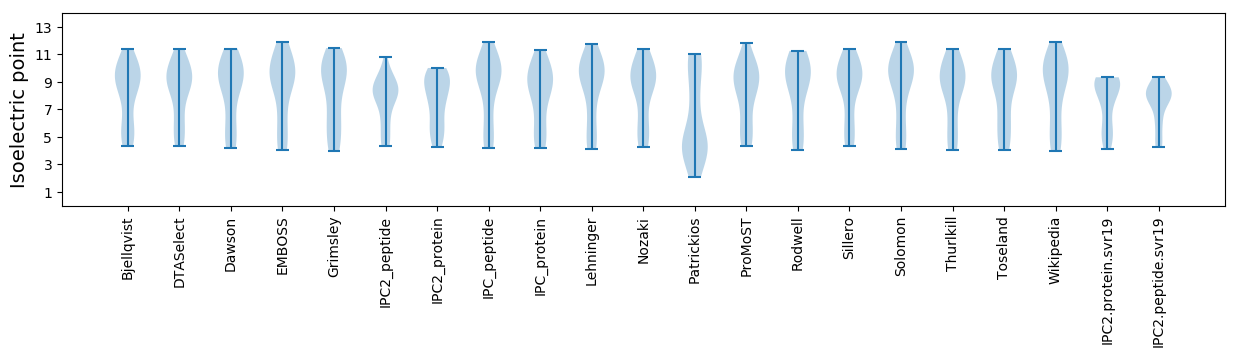
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C8CHL2|C8CHL2_9VIRU Uncharacterized protein OS=Thermus virus P23-77 OX=1714272 PE=4 SV=1
MM1 pKa = 7.85GGRR4 pKa = 11.84QTFRR8 pKa = 11.84LLIIGKK14 pKa = 9.17SGSGKK19 pKa = 8.61STLARR24 pKa = 11.84QVIRR28 pKa = 11.84AMEE31 pKa = 3.88GRR33 pKa = 11.84YY34 pKa = 8.96RR35 pKa = 11.84HH36 pKa = 5.78LVIVNRR42 pKa = 11.84KK43 pKa = 6.84TEE45 pKa = 3.93FAEE48 pKa = 3.97LAEE51 pKa = 4.04GRR53 pKa = 11.84FRR55 pKa = 11.84VKK57 pKa = 10.64EE58 pKa = 4.38DD59 pKa = 3.56GDD61 pKa = 3.85PGPALRR67 pKa = 11.84RR68 pKa = 11.84YY69 pKa = 9.59RR70 pKa = 11.84RR71 pKa = 11.84VHH73 pKa = 5.0FHH75 pKa = 4.4VTGYY79 pKa = 10.74DD80 pKa = 3.33PRR82 pKa = 11.84PFLDD86 pKa = 4.01ALGQEE91 pKa = 4.79LMRR94 pKa = 11.84MRR96 pKa = 11.84DD97 pKa = 3.26VLLVVDD103 pKa = 5.02EE104 pKa = 4.33AHH106 pKa = 6.4QFFPRR111 pKa = 11.84GQVPKK116 pKa = 10.89GLFEE120 pKa = 4.2VLTGGRR126 pKa = 11.84EE127 pKa = 3.82NGHH130 pKa = 5.49NVVFVTQMLKK140 pKa = 10.75GAVGGIDD147 pKa = 3.08PGVRR151 pKa = 11.84RR152 pKa = 11.84QASHH156 pKa = 6.64LVAFRR161 pKa = 11.84VTEE164 pKa = 4.02PAEE167 pKa = 4.15VASVAEE173 pKa = 4.34MFPEE177 pKa = 4.3LGEE180 pKa = 4.24RR181 pKa = 11.84VQHH184 pKa = 6.03LRR186 pKa = 11.84RR187 pKa = 11.84PEE189 pKa = 4.26GGLPPEE195 pKa = 4.36YY196 pKa = 10.36GVRR199 pKa = 11.84DD200 pKa = 3.75LDD202 pKa = 3.72RR203 pKa = 11.84DD204 pKa = 3.42RR205 pKa = 11.84SGLVLRR211 pKa = 11.84DD212 pKa = 3.19PRR214 pKa = 11.84DD215 pKa = 3.11PRR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84VWVPLL224 pKa = 3.52
MM1 pKa = 7.85GGRR4 pKa = 11.84QTFRR8 pKa = 11.84LLIIGKK14 pKa = 9.17SGSGKK19 pKa = 8.61STLARR24 pKa = 11.84QVIRR28 pKa = 11.84AMEE31 pKa = 3.88GRR33 pKa = 11.84YY34 pKa = 8.96RR35 pKa = 11.84HH36 pKa = 5.78LVIVNRR42 pKa = 11.84KK43 pKa = 6.84TEE45 pKa = 3.93FAEE48 pKa = 3.97LAEE51 pKa = 4.04GRR53 pKa = 11.84FRR55 pKa = 11.84VKK57 pKa = 10.64EE58 pKa = 4.38DD59 pKa = 3.56GDD61 pKa = 3.85PGPALRR67 pKa = 11.84RR68 pKa = 11.84YY69 pKa = 9.59RR70 pKa = 11.84RR71 pKa = 11.84VHH73 pKa = 5.0FHH75 pKa = 4.4VTGYY79 pKa = 10.74DD80 pKa = 3.33PRR82 pKa = 11.84PFLDD86 pKa = 4.01ALGQEE91 pKa = 4.79LMRR94 pKa = 11.84MRR96 pKa = 11.84DD97 pKa = 3.26VLLVVDD103 pKa = 5.02EE104 pKa = 4.33AHH106 pKa = 6.4QFFPRR111 pKa = 11.84GQVPKK116 pKa = 10.89GLFEE120 pKa = 4.2VLTGGRR126 pKa = 11.84EE127 pKa = 3.82NGHH130 pKa = 5.49NVVFVTQMLKK140 pKa = 10.75GAVGGIDD147 pKa = 3.08PGVRR151 pKa = 11.84RR152 pKa = 11.84QASHH156 pKa = 6.64LVAFRR161 pKa = 11.84VTEE164 pKa = 4.02PAEE167 pKa = 4.15VASVAEE173 pKa = 4.34MFPEE177 pKa = 4.3LGEE180 pKa = 4.24RR181 pKa = 11.84VQHH184 pKa = 6.03LRR186 pKa = 11.84RR187 pKa = 11.84PEE189 pKa = 4.26GGLPPEE195 pKa = 4.36YY196 pKa = 10.36GVRR199 pKa = 11.84DD200 pKa = 3.75LDD202 pKa = 3.72RR203 pKa = 11.84DD204 pKa = 3.42RR205 pKa = 11.84SGLVLRR211 pKa = 11.84DD212 pKa = 3.19PRR214 pKa = 11.84DD215 pKa = 3.11PRR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84VWVPLL224 pKa = 3.52
Molecular weight: 25.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5621 |
35 |
425 |
151.9 |
16.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.119 ± 0.663 | 0.747 ± 0.179 |
3.896 ± 0.256 | 5.675 ± 0.428 |
2.597 ± 0.267 | 9.998 ± 0.481 |
1.228 ± 0.155 | 3.434 ± 0.304 |
3.096 ± 0.474 | 10.532 ± 0.433 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.975 ± 0.146 | 1.939 ± 0.287 |
8.023 ± 0.511 | 4.341 ± 0.272 |
8.308 ± 0.692 | 4.892 ± 0.342 |
4.892 ± 0.461 | 8.308 ± 0.471 |
2.028 ± 0.207 | 2.971 ± 0.323 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
