
Sphingobium fluviale
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingobium
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
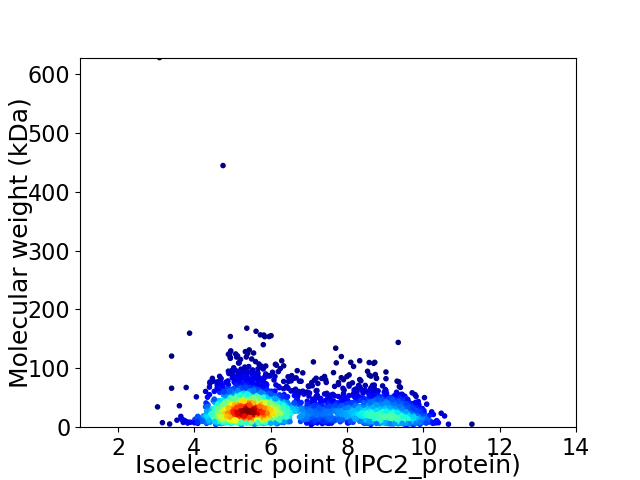
Virtual 2D-PAGE plot for 2932 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A4Q1KID1|A0A4Q1KID1_9SPHN NAD(P)H-quinone oxidoreductase OS=Sphingobium fluviale OX=2506423 GN=EQG66_08315 PE=4 SV=1
MM1 pKa = 7.66AIEE4 pKa = 4.35PNSPLFQAILAMDD17 pKa = 4.27SYY19 pKa = 11.86SRR21 pKa = 11.84GYY23 pKa = 10.82LSGVDD28 pKa = 3.31LRR30 pKa = 11.84LRR32 pKa = 11.84EE33 pKa = 4.21EE34 pKa = 4.17NGNLVVDD41 pKa = 4.46EE42 pKa = 4.88NGAPIPSDD50 pKa = 3.46TIGNTLGHH58 pKa = 6.23LTIIANSSEE67 pKa = 4.21AFPSGDD73 pKa = 3.4DD74 pKa = 3.51QKK76 pKa = 11.87AGFYY80 pKa = 10.47AIAYY84 pKa = 8.62QYY86 pKa = 10.99TDD88 pKa = 3.62ANSQIKK94 pKa = 9.1TIISYY99 pKa = 10.61RR100 pKa = 11.84GTDD103 pKa = 3.42QFFTTYY109 pKa = 10.74DD110 pKa = 3.33ANNNPLVGGDD120 pKa = 3.17IANAYY125 pKa = 9.91LVGSGAAEE133 pKa = 3.68QTQARR138 pKa = 11.84LAIEE142 pKa = 5.02FYY144 pKa = 10.76KK145 pKa = 10.9SVAAAINPQNPDD157 pKa = 3.02PYY159 pKa = 10.49AANIVVTGHH168 pKa = 5.98SSGGGLAGLVGAIYY182 pKa = 10.14HH183 pKa = 6.19KK184 pKa = 10.71EE185 pKa = 3.71GVVFDD190 pKa = 3.78NMAFVEE196 pKa = 4.3AARR199 pKa = 11.84NLSFYY204 pKa = 10.44NGSPSSPNDD213 pKa = 3.28PSDD216 pKa = 3.79LLRR219 pKa = 11.84PLYY222 pKa = 10.94SEE224 pKa = 6.07DD225 pKa = 4.07FYY227 pKa = 11.9QLINGTNGPVLIDD240 pKa = 3.67GNGQRR245 pKa = 11.84SLDD248 pKa = 3.85PRR250 pKa = 11.84WQSGGDD256 pKa = 3.64FYY258 pKa = 11.97AKK260 pKa = 10.57FEE262 pKa = 4.08GLKK265 pKa = 10.14AYY267 pKa = 8.7TIAGEE272 pKa = 3.94ILAEE276 pKa = 4.09DD277 pKa = 5.36FIPGGNAHH285 pKa = 6.59QDD287 pKa = 3.2PVKK290 pKa = 10.33PVEE293 pKa = 4.28LSLAADD299 pKa = 3.54VGWFNLIGDD308 pKa = 3.98NGIDD312 pKa = 3.7PFDD315 pKa = 3.63AHH317 pKa = 6.52NVATLVIRR325 pKa = 11.84MFAGITPAEE334 pKa = 4.05EE335 pKa = 3.87AAFGHH340 pKa = 6.3AWKK343 pKa = 10.2IAAPHH348 pKa = 6.38FWPVMYY354 pKa = 10.32DD355 pKa = 3.38NQFSQSIGVGVADD368 pKa = 4.34GGRR371 pKa = 11.84YY372 pKa = 8.43IAEE375 pKa = 4.61NGPHH379 pKa = 7.03GYY381 pKa = 10.42DD382 pKa = 2.78KK383 pKa = 11.32TMRR386 pKa = 11.84TYY388 pKa = 10.47IAYY391 pKa = 10.18SAIDD395 pKa = 3.64NGADD399 pKa = 3.47DD400 pKa = 5.39LSARR404 pKa = 11.84PYY406 pKa = 10.82GDD408 pKa = 3.11TAIRR412 pKa = 11.84ALYY415 pKa = 10.53DD416 pKa = 3.63DD417 pKa = 5.32ASDD420 pKa = 4.12LGVAMGVASKK430 pKa = 10.72GAAVLRR436 pKa = 11.84YY437 pKa = 9.64AEE439 pKa = 4.1QLSKK443 pKa = 11.41VFVEE447 pKa = 5.87FAAKK451 pKa = 10.14LAMEE455 pKa = 4.79KK456 pKa = 10.36VLQSSDD462 pKa = 3.16YY463 pKa = 9.46ATEE466 pKa = 4.06HH467 pKa = 6.57NALQGILSLTEE478 pKa = 4.09GNTGITADD486 pKa = 4.13FSSAKK491 pKa = 9.1WQFDD495 pKa = 3.26NEE497 pKa = 4.22APSNIISKK505 pKa = 10.1QLLLGNLRR513 pKa = 11.84IDD515 pKa = 4.65DD516 pKa = 4.15LQKK519 pKa = 11.01QYY521 pKa = 11.33ILDD524 pKa = 3.79QFEE527 pKa = 5.21HH528 pKa = 5.82IQKK531 pKa = 8.38ITISTTRR538 pKa = 11.84DD539 pKa = 2.8AFNYY543 pKa = 8.35VIPASGGNSTQMFIEE558 pKa = 4.33NSTSEE563 pKa = 4.46STSGAQIVVGGAGNDD578 pKa = 3.1AVYY581 pKa = 11.04ARR583 pKa = 11.84IGGDD587 pKa = 3.44FVIGGHH593 pKa = 6.99GIDD596 pKa = 3.38TVDD599 pKa = 3.16YY600 pKa = 9.59SGIYY604 pKa = 8.84IAPPEE609 pKa = 4.31GEE611 pKa = 4.73IYY613 pKa = 9.37PAPNYY618 pKa = 10.41DD619 pKa = 3.61PYY621 pKa = 11.6SYY623 pKa = 8.08TTGLEE628 pKa = 3.97ADD630 pKa = 5.17FSAAAEE636 pKa = 4.37SGFGTVKK643 pKa = 10.76LMGSMHH649 pKa = 5.5TVKK652 pKa = 10.59DD653 pKa = 3.55YY654 pKa = 11.37LFSVEE659 pKa = 5.26HH660 pKa = 6.08LTLTDD665 pKa = 3.61YY666 pKa = 11.76DD667 pKa = 4.14DD668 pKa = 5.29GLWLSTPVAGVEE680 pKa = 3.74RR681 pKa = 11.84WFDD684 pKa = 3.26GGAGGDD690 pKa = 3.52TVFYY694 pKa = 8.89DD695 pKa = 3.72TSNLIRR701 pKa = 11.84DD702 pKa = 4.07DD703 pKa = 4.12VNSVVQNINGTGKK716 pKa = 10.16DD717 pKa = 3.14HH718 pKa = 7.07LLNVDD723 pKa = 4.71FFGFSPSQYY732 pKa = 9.99YY733 pKa = 10.66DD734 pKa = 2.93IVNGNLPGPVLNMAAKK750 pKa = 10.26YY751 pKa = 9.69IIPDD755 pKa = 2.94VRR757 pKa = 11.84YY758 pKa = 7.74TYY760 pKa = 10.99DD761 pKa = 3.24VNASSASTLDD771 pKa = 3.62YY772 pKa = 10.23STSAANILFTLGVQYY787 pKa = 9.46NTAEE791 pKa = 4.16MFDD794 pKa = 3.35ASGVHH799 pKa = 4.98THH801 pKa = 7.43LISEE805 pKa = 4.5TPSMVIGSNAVGNTISFYY823 pKa = 10.42GTGAFTGGIQDD834 pKa = 3.79DD835 pKa = 4.64TIVIQTGGFEE845 pKa = 4.23TMGRR849 pKa = 11.84ITIGYY854 pKa = 8.42QGGYY858 pKa = 9.04DD859 pKa = 3.42QIVSYY864 pKa = 10.8GSEE867 pKa = 4.18LAPGLNPVNAPLLILMANGILPDD890 pKa = 3.56QVSVEE895 pKa = 4.09FFNQSEE901 pKa = 4.21QDD903 pKa = 3.28GDD905 pKa = 4.24PLVGTYY911 pKa = 9.98FPVTYY916 pKa = 10.4ADD918 pKa = 3.85ALITVDD924 pKa = 4.06GYY926 pKa = 10.26GTISIEE932 pKa = 4.52GLKK935 pKa = 10.44LYY937 pKa = 10.9NNGSFVPLQIAFGDD951 pKa = 3.87GTTWEE956 pKa = 4.25WGEE959 pKa = 4.21TGQPASLFDD968 pKa = 3.98YY969 pKa = 7.92DD970 pKa = 3.66TSFFGKK976 pKa = 10.64GNWDD980 pKa = 3.52NNVLTGYY987 pKa = 9.79EE988 pKa = 4.23GQDD991 pKa = 3.25NTLHH995 pKa = 6.66GFGGDD1000 pKa = 3.49DD1001 pKa = 4.26TIIASYY1007 pKa = 11.36GNDD1010 pKa = 3.17LVYY1013 pKa = 10.97GGDD1016 pKa = 3.69GEE1018 pKa = 5.13DD1019 pKa = 5.12FIDD1022 pKa = 5.02AGMGVNTVYY1031 pKa = 10.95GGFGADD1037 pKa = 3.02TVVMSPYY1044 pKa = 10.63SVTNFVDD1051 pKa = 3.43YY1052 pKa = 9.15TAAQKK1057 pKa = 10.67DD1058 pKa = 4.34IIILPYY1064 pKa = 8.81TVSDD1068 pKa = 3.59ISQLVTSWWTSPHH1081 pKa = 6.06GAQGLEE1087 pKa = 4.12LYY1089 pKa = 10.83ADD1091 pKa = 3.92NTSMLLGIEE1100 pKa = 4.03RR1101 pKa = 11.84PEE1103 pKa = 5.33AEE1105 pKa = 5.16DD1106 pKa = 3.08EE1107 pKa = 4.34DD1108 pKa = 4.41AVVRR1112 pKa = 11.84VQLNSGTKK1120 pKa = 10.26LSLAVDD1126 pKa = 3.82RR1127 pKa = 11.84DD1128 pKa = 3.8GLITSVTSIGSGGDD1142 pKa = 3.14DD1143 pKa = 3.58DD1144 pKa = 6.29GIYY1147 pKa = 8.25GTPDD1151 pKa = 3.18NDD1153 pKa = 4.32TLHH1156 pKa = 5.38GTAWRR1161 pKa = 11.84DD1162 pKa = 3.67FIYY1165 pKa = 10.82GLAGDD1170 pKa = 4.13DD1171 pKa = 4.5VIWGYY1176 pKa = 11.78ADD1178 pKa = 4.65DD1179 pKa = 4.62DD1180 pKa = 5.21VIVGGAGDD1188 pKa = 3.63DD1189 pKa = 3.63TLYY1192 pKa = 11.4GGVGNDD1198 pKa = 3.34NYY1200 pKa = 10.49WFNLGDD1206 pKa = 3.92GSDD1209 pKa = 3.96TIVDD1213 pKa = 3.62EE1214 pKa = 4.92GGSLDD1219 pKa = 4.09RR1220 pKa = 11.84IRR1222 pKa = 11.84FAASVEE1228 pKa = 4.3SSSVSYY1234 pKa = 10.79YY1235 pKa = 10.59QDD1237 pKa = 3.27GDD1239 pKa = 3.99DD1240 pKa = 4.3LFIRR1244 pKa = 11.84YY1245 pKa = 8.09GAGSDD1250 pKa = 3.84EE1251 pKa = 4.05ITVTGFFASDD1261 pKa = 3.11EE1262 pKa = 4.27TRR1264 pKa = 11.84IEE1266 pKa = 3.87EE1267 pKa = 4.19VAFMDD1272 pKa = 4.83GTIHH1276 pKa = 7.2DD1277 pKa = 4.06AAYY1280 pKa = 9.86ILAAVSTGTGTITGTSGADD1299 pKa = 3.27LLEE1302 pKa = 4.49GTEE1305 pKa = 4.06AADD1308 pKa = 3.43ILRR1311 pKa = 11.84GLGGADD1317 pKa = 2.98TLYY1320 pKa = 10.91GYY1322 pKa = 10.78DD1323 pKa = 4.53GDD1325 pKa = 5.84DD1326 pKa = 3.51ILIGGNGHH1334 pKa = 7.05DD1335 pKa = 4.2SLYY1338 pKa = 11.25GGIGNDD1344 pKa = 3.08ILRR1347 pKa = 11.84GGSGNDD1353 pKa = 3.6LLIGGAGNDD1362 pKa = 3.47ILKK1365 pKa = 10.74GSAGNDD1371 pKa = 2.98ILRR1374 pKa = 11.84GGSGDD1379 pKa = 4.15DD1380 pKa = 3.27ILRR1383 pKa = 11.84GGDD1386 pKa = 3.33GDD1388 pKa = 4.0DD1389 pKa = 3.39RR1390 pKa = 11.84LVGGRR1395 pKa = 11.84GSDD1398 pKa = 3.4TLDD1401 pKa = 3.49GGNGVDD1407 pKa = 3.23TFVFRR1412 pKa = 11.84ANHH1415 pKa = 6.3IGTGVDD1421 pKa = 3.61TIKK1424 pKa = 11.02DD1425 pKa = 3.46FSLAQNDD1432 pKa = 4.6KK1433 pKa = 10.86IDD1435 pKa = 3.84ISDD1438 pKa = 4.31LLSAYY1443 pKa = 10.27DD1444 pKa = 4.13PLTDD1448 pKa = 3.76ALADD1452 pKa = 3.95FVQFTRR1458 pKa = 11.84SGKK1461 pKa = 8.99KK1462 pKa = 9.86SNMFVDD1468 pKa = 4.23LDD1470 pKa = 4.04GAGAEE1475 pKa = 4.3HH1476 pKa = 7.29DD1477 pKa = 3.87WVHH1480 pKa = 5.63VATLQGVTALPDD1492 pKa = 3.25VDD1494 pKa = 3.67TLVANGHH1501 pKa = 6.54LLAAA1505 pKa = 5.26
MM1 pKa = 7.66AIEE4 pKa = 4.35PNSPLFQAILAMDD17 pKa = 4.27SYY19 pKa = 11.86SRR21 pKa = 11.84GYY23 pKa = 10.82LSGVDD28 pKa = 3.31LRR30 pKa = 11.84LRR32 pKa = 11.84EE33 pKa = 4.21EE34 pKa = 4.17NGNLVVDD41 pKa = 4.46EE42 pKa = 4.88NGAPIPSDD50 pKa = 3.46TIGNTLGHH58 pKa = 6.23LTIIANSSEE67 pKa = 4.21AFPSGDD73 pKa = 3.4DD74 pKa = 3.51QKK76 pKa = 11.87AGFYY80 pKa = 10.47AIAYY84 pKa = 8.62QYY86 pKa = 10.99TDD88 pKa = 3.62ANSQIKK94 pKa = 9.1TIISYY99 pKa = 10.61RR100 pKa = 11.84GTDD103 pKa = 3.42QFFTTYY109 pKa = 10.74DD110 pKa = 3.33ANNNPLVGGDD120 pKa = 3.17IANAYY125 pKa = 9.91LVGSGAAEE133 pKa = 3.68QTQARR138 pKa = 11.84LAIEE142 pKa = 5.02FYY144 pKa = 10.76KK145 pKa = 10.9SVAAAINPQNPDD157 pKa = 3.02PYY159 pKa = 10.49AANIVVTGHH168 pKa = 5.98SSGGGLAGLVGAIYY182 pKa = 10.14HH183 pKa = 6.19KK184 pKa = 10.71EE185 pKa = 3.71GVVFDD190 pKa = 3.78NMAFVEE196 pKa = 4.3AARR199 pKa = 11.84NLSFYY204 pKa = 10.44NGSPSSPNDD213 pKa = 3.28PSDD216 pKa = 3.79LLRR219 pKa = 11.84PLYY222 pKa = 10.94SEE224 pKa = 6.07DD225 pKa = 4.07FYY227 pKa = 11.9QLINGTNGPVLIDD240 pKa = 3.67GNGQRR245 pKa = 11.84SLDD248 pKa = 3.85PRR250 pKa = 11.84WQSGGDD256 pKa = 3.64FYY258 pKa = 11.97AKK260 pKa = 10.57FEE262 pKa = 4.08GLKK265 pKa = 10.14AYY267 pKa = 8.7TIAGEE272 pKa = 3.94ILAEE276 pKa = 4.09DD277 pKa = 5.36FIPGGNAHH285 pKa = 6.59QDD287 pKa = 3.2PVKK290 pKa = 10.33PVEE293 pKa = 4.28LSLAADD299 pKa = 3.54VGWFNLIGDD308 pKa = 3.98NGIDD312 pKa = 3.7PFDD315 pKa = 3.63AHH317 pKa = 6.52NVATLVIRR325 pKa = 11.84MFAGITPAEE334 pKa = 4.05EE335 pKa = 3.87AAFGHH340 pKa = 6.3AWKK343 pKa = 10.2IAAPHH348 pKa = 6.38FWPVMYY354 pKa = 10.32DD355 pKa = 3.38NQFSQSIGVGVADD368 pKa = 4.34GGRR371 pKa = 11.84YY372 pKa = 8.43IAEE375 pKa = 4.61NGPHH379 pKa = 7.03GYY381 pKa = 10.42DD382 pKa = 2.78KK383 pKa = 11.32TMRR386 pKa = 11.84TYY388 pKa = 10.47IAYY391 pKa = 10.18SAIDD395 pKa = 3.64NGADD399 pKa = 3.47DD400 pKa = 5.39LSARR404 pKa = 11.84PYY406 pKa = 10.82GDD408 pKa = 3.11TAIRR412 pKa = 11.84ALYY415 pKa = 10.53DD416 pKa = 3.63DD417 pKa = 5.32ASDD420 pKa = 4.12LGVAMGVASKK430 pKa = 10.72GAAVLRR436 pKa = 11.84YY437 pKa = 9.64AEE439 pKa = 4.1QLSKK443 pKa = 11.41VFVEE447 pKa = 5.87FAAKK451 pKa = 10.14LAMEE455 pKa = 4.79KK456 pKa = 10.36VLQSSDD462 pKa = 3.16YY463 pKa = 9.46ATEE466 pKa = 4.06HH467 pKa = 6.57NALQGILSLTEE478 pKa = 4.09GNTGITADD486 pKa = 4.13FSSAKK491 pKa = 9.1WQFDD495 pKa = 3.26NEE497 pKa = 4.22APSNIISKK505 pKa = 10.1QLLLGNLRR513 pKa = 11.84IDD515 pKa = 4.65DD516 pKa = 4.15LQKK519 pKa = 11.01QYY521 pKa = 11.33ILDD524 pKa = 3.79QFEE527 pKa = 5.21HH528 pKa = 5.82IQKK531 pKa = 8.38ITISTTRR538 pKa = 11.84DD539 pKa = 2.8AFNYY543 pKa = 8.35VIPASGGNSTQMFIEE558 pKa = 4.33NSTSEE563 pKa = 4.46STSGAQIVVGGAGNDD578 pKa = 3.1AVYY581 pKa = 11.04ARR583 pKa = 11.84IGGDD587 pKa = 3.44FVIGGHH593 pKa = 6.99GIDD596 pKa = 3.38TVDD599 pKa = 3.16YY600 pKa = 9.59SGIYY604 pKa = 8.84IAPPEE609 pKa = 4.31GEE611 pKa = 4.73IYY613 pKa = 9.37PAPNYY618 pKa = 10.41DD619 pKa = 3.61PYY621 pKa = 11.6SYY623 pKa = 8.08TTGLEE628 pKa = 3.97ADD630 pKa = 5.17FSAAAEE636 pKa = 4.37SGFGTVKK643 pKa = 10.76LMGSMHH649 pKa = 5.5TVKK652 pKa = 10.59DD653 pKa = 3.55YY654 pKa = 11.37LFSVEE659 pKa = 5.26HH660 pKa = 6.08LTLTDD665 pKa = 3.61YY666 pKa = 11.76DD667 pKa = 4.14DD668 pKa = 5.29GLWLSTPVAGVEE680 pKa = 3.74RR681 pKa = 11.84WFDD684 pKa = 3.26GGAGGDD690 pKa = 3.52TVFYY694 pKa = 8.89DD695 pKa = 3.72TSNLIRR701 pKa = 11.84DD702 pKa = 4.07DD703 pKa = 4.12VNSVVQNINGTGKK716 pKa = 10.16DD717 pKa = 3.14HH718 pKa = 7.07LLNVDD723 pKa = 4.71FFGFSPSQYY732 pKa = 9.99YY733 pKa = 10.66DD734 pKa = 2.93IVNGNLPGPVLNMAAKK750 pKa = 10.26YY751 pKa = 9.69IIPDD755 pKa = 2.94VRR757 pKa = 11.84YY758 pKa = 7.74TYY760 pKa = 10.99DD761 pKa = 3.24VNASSASTLDD771 pKa = 3.62YY772 pKa = 10.23STSAANILFTLGVQYY787 pKa = 9.46NTAEE791 pKa = 4.16MFDD794 pKa = 3.35ASGVHH799 pKa = 4.98THH801 pKa = 7.43LISEE805 pKa = 4.5TPSMVIGSNAVGNTISFYY823 pKa = 10.42GTGAFTGGIQDD834 pKa = 3.79DD835 pKa = 4.64TIVIQTGGFEE845 pKa = 4.23TMGRR849 pKa = 11.84ITIGYY854 pKa = 8.42QGGYY858 pKa = 9.04DD859 pKa = 3.42QIVSYY864 pKa = 10.8GSEE867 pKa = 4.18LAPGLNPVNAPLLILMANGILPDD890 pKa = 3.56QVSVEE895 pKa = 4.09FFNQSEE901 pKa = 4.21QDD903 pKa = 3.28GDD905 pKa = 4.24PLVGTYY911 pKa = 9.98FPVTYY916 pKa = 10.4ADD918 pKa = 3.85ALITVDD924 pKa = 4.06GYY926 pKa = 10.26GTISIEE932 pKa = 4.52GLKK935 pKa = 10.44LYY937 pKa = 10.9NNGSFVPLQIAFGDD951 pKa = 3.87GTTWEE956 pKa = 4.25WGEE959 pKa = 4.21TGQPASLFDD968 pKa = 3.98YY969 pKa = 7.92DD970 pKa = 3.66TSFFGKK976 pKa = 10.64GNWDD980 pKa = 3.52NNVLTGYY987 pKa = 9.79EE988 pKa = 4.23GQDD991 pKa = 3.25NTLHH995 pKa = 6.66GFGGDD1000 pKa = 3.49DD1001 pKa = 4.26TIIASYY1007 pKa = 11.36GNDD1010 pKa = 3.17LVYY1013 pKa = 10.97GGDD1016 pKa = 3.69GEE1018 pKa = 5.13DD1019 pKa = 5.12FIDD1022 pKa = 5.02AGMGVNTVYY1031 pKa = 10.95GGFGADD1037 pKa = 3.02TVVMSPYY1044 pKa = 10.63SVTNFVDD1051 pKa = 3.43YY1052 pKa = 9.15TAAQKK1057 pKa = 10.67DD1058 pKa = 4.34IIILPYY1064 pKa = 8.81TVSDD1068 pKa = 3.59ISQLVTSWWTSPHH1081 pKa = 6.06GAQGLEE1087 pKa = 4.12LYY1089 pKa = 10.83ADD1091 pKa = 3.92NTSMLLGIEE1100 pKa = 4.03RR1101 pKa = 11.84PEE1103 pKa = 5.33AEE1105 pKa = 5.16DD1106 pKa = 3.08EE1107 pKa = 4.34DD1108 pKa = 4.41AVVRR1112 pKa = 11.84VQLNSGTKK1120 pKa = 10.26LSLAVDD1126 pKa = 3.82RR1127 pKa = 11.84DD1128 pKa = 3.8GLITSVTSIGSGGDD1142 pKa = 3.14DD1143 pKa = 3.58DD1144 pKa = 6.29GIYY1147 pKa = 8.25GTPDD1151 pKa = 3.18NDD1153 pKa = 4.32TLHH1156 pKa = 5.38GTAWRR1161 pKa = 11.84DD1162 pKa = 3.67FIYY1165 pKa = 10.82GLAGDD1170 pKa = 4.13DD1171 pKa = 4.5VIWGYY1176 pKa = 11.78ADD1178 pKa = 4.65DD1179 pKa = 4.62DD1180 pKa = 5.21VIVGGAGDD1188 pKa = 3.63DD1189 pKa = 3.63TLYY1192 pKa = 11.4GGVGNDD1198 pKa = 3.34NYY1200 pKa = 10.49WFNLGDD1206 pKa = 3.92GSDD1209 pKa = 3.96TIVDD1213 pKa = 3.62EE1214 pKa = 4.92GGSLDD1219 pKa = 4.09RR1220 pKa = 11.84IRR1222 pKa = 11.84FAASVEE1228 pKa = 4.3SSSVSYY1234 pKa = 10.79YY1235 pKa = 10.59QDD1237 pKa = 3.27GDD1239 pKa = 3.99DD1240 pKa = 4.3LFIRR1244 pKa = 11.84YY1245 pKa = 8.09GAGSDD1250 pKa = 3.84EE1251 pKa = 4.05ITVTGFFASDD1261 pKa = 3.11EE1262 pKa = 4.27TRR1264 pKa = 11.84IEE1266 pKa = 3.87EE1267 pKa = 4.19VAFMDD1272 pKa = 4.83GTIHH1276 pKa = 7.2DD1277 pKa = 4.06AAYY1280 pKa = 9.86ILAAVSTGTGTITGTSGADD1299 pKa = 3.27LLEE1302 pKa = 4.49GTEE1305 pKa = 4.06AADD1308 pKa = 3.43ILRR1311 pKa = 11.84GLGGADD1317 pKa = 2.98TLYY1320 pKa = 10.91GYY1322 pKa = 10.78DD1323 pKa = 4.53GDD1325 pKa = 5.84DD1326 pKa = 3.51ILIGGNGHH1334 pKa = 7.05DD1335 pKa = 4.2SLYY1338 pKa = 11.25GGIGNDD1344 pKa = 3.08ILRR1347 pKa = 11.84GGSGNDD1353 pKa = 3.6LLIGGAGNDD1362 pKa = 3.47ILKK1365 pKa = 10.74GSAGNDD1371 pKa = 2.98ILRR1374 pKa = 11.84GGSGDD1379 pKa = 4.15DD1380 pKa = 3.27ILRR1383 pKa = 11.84GGDD1386 pKa = 3.33GDD1388 pKa = 4.0DD1389 pKa = 3.39RR1390 pKa = 11.84LVGGRR1395 pKa = 11.84GSDD1398 pKa = 3.4TLDD1401 pKa = 3.49GGNGVDD1407 pKa = 3.23TFVFRR1412 pKa = 11.84ANHH1415 pKa = 6.3IGTGVDD1421 pKa = 3.61TIKK1424 pKa = 11.02DD1425 pKa = 3.46FSLAQNDD1432 pKa = 4.6KK1433 pKa = 10.86IDD1435 pKa = 3.84ISDD1438 pKa = 4.31LLSAYY1443 pKa = 10.27DD1444 pKa = 4.13PLTDD1448 pKa = 3.76ALADD1452 pKa = 3.95FVQFTRR1458 pKa = 11.84SGKK1461 pKa = 8.99KK1462 pKa = 9.86SNMFVDD1468 pKa = 4.23LDD1470 pKa = 4.04GAGAEE1475 pKa = 4.3HH1476 pKa = 7.29DD1477 pKa = 3.87WVHH1480 pKa = 5.63VATLQGVTALPDD1492 pKa = 3.25VDD1494 pKa = 3.67TLVANGHH1501 pKa = 6.54LLAAA1505 pKa = 5.26
Molecular weight: 159.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q1KGR2|A0A4Q1KGR2_9SPHN UvrABC system protein C OS=Sphingobium fluviale OX=2506423 GN=uvrC PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84NILRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84NILRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
915520 |
23 |
6626 |
312.3 |
33.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.108 ± 0.066 | 0.873 ± 0.017 |
5.874 ± 0.055 | 5.559 ± 0.048 |
3.405 ± 0.032 | 8.755 ± 0.104 |
2.073 ± 0.026 | 5.364 ± 0.028 |
3.296 ± 0.041 | 9.917 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.618 ± 0.026 | 2.659 ± 0.035 |
5.074 ± 0.044 | 3.206 ± 0.027 |
7.082 ± 0.063 | 5.527 ± 0.047 |
5.114 ± 0.051 | 6.908 ± 0.037 |
1.366 ± 0.019 | 2.223 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
