
Streptococcus satellite phage Javan631
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
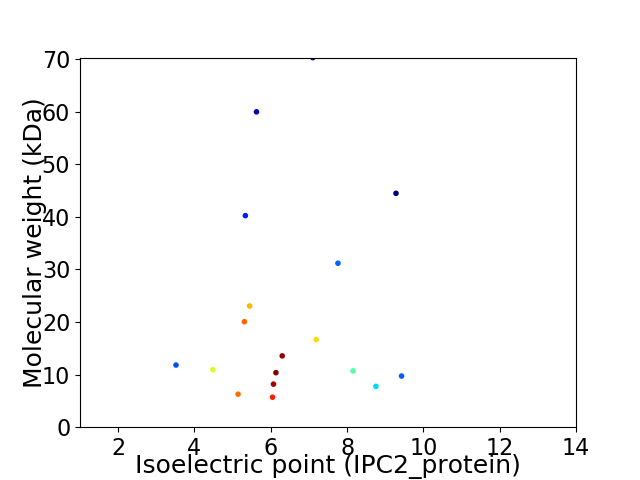
Virtual 2D-PAGE plot for 18 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZSN9|A0A4D5ZSN9_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan631 OX=2558795 GN=JavanS631_0007 PE=4 SV=1
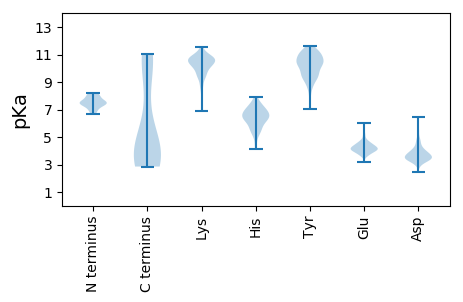
MM1 pKa = 7.6KK2 pKa = 10.4NLEE5 pKa = 4.17EE6 pKa = 5.44KK7 pKa = 10.32IEE9 pKa = 4.01PVVIILDD16 pKa = 3.82EE17 pKa = 4.39EE18 pKa = 4.86DD19 pKa = 5.34DD20 pKa = 4.57FDD22 pKa = 5.98FEE24 pKa = 4.44PTDD27 pKa = 4.67EE28 pKa = 4.34EE29 pKa = 5.21LQDD32 pKa = 3.54QYY34 pKa = 11.6EE35 pKa = 4.32AEE37 pKa = 4.3CLEE40 pKa = 5.78DD41 pKa = 3.71DD42 pKa = 4.11TQDD45 pKa = 3.43VINWINEE52 pKa = 4.11SNTLAEE58 pKa = 4.29YY59 pKa = 11.21VSMEE63 pKa = 4.26FEE65 pKa = 4.68NMNKK69 pKa = 10.07DD70 pKa = 3.67DD71 pKa = 5.63LGTSLEE77 pKa = 4.21NASQCLDD84 pKa = 4.31AIVGMLEE91 pKa = 3.77NGTLILAGGKK101 pKa = 9.32HH102 pKa = 5.43EE103 pKa = 4.13NN104 pKa = 3.61
MM1 pKa = 7.6KK2 pKa = 10.4NLEE5 pKa = 4.17EE6 pKa = 5.44KK7 pKa = 10.32IEE9 pKa = 4.01PVVIILDD16 pKa = 3.82EE17 pKa = 4.39EE18 pKa = 4.86DD19 pKa = 5.34DD20 pKa = 4.57FDD22 pKa = 5.98FEE24 pKa = 4.44PTDD27 pKa = 4.67EE28 pKa = 4.34EE29 pKa = 5.21LQDD32 pKa = 3.54QYY34 pKa = 11.6EE35 pKa = 4.32AEE37 pKa = 4.3CLEE40 pKa = 5.78DD41 pKa = 3.71DD42 pKa = 4.11TQDD45 pKa = 3.43VINWINEE52 pKa = 4.11SNTLAEE58 pKa = 4.29YY59 pKa = 11.21VSMEE63 pKa = 4.26FEE65 pKa = 4.68NMNKK69 pKa = 10.07DD70 pKa = 3.67DD71 pKa = 5.63LGTSLEE77 pKa = 4.21NASQCLDD84 pKa = 4.31AIVGMLEE91 pKa = 3.77NGTLILAGGKK101 pKa = 9.32HH102 pKa = 5.43EE103 pKa = 4.13NN104 pKa = 3.61
Molecular weight: 11.82 kDa
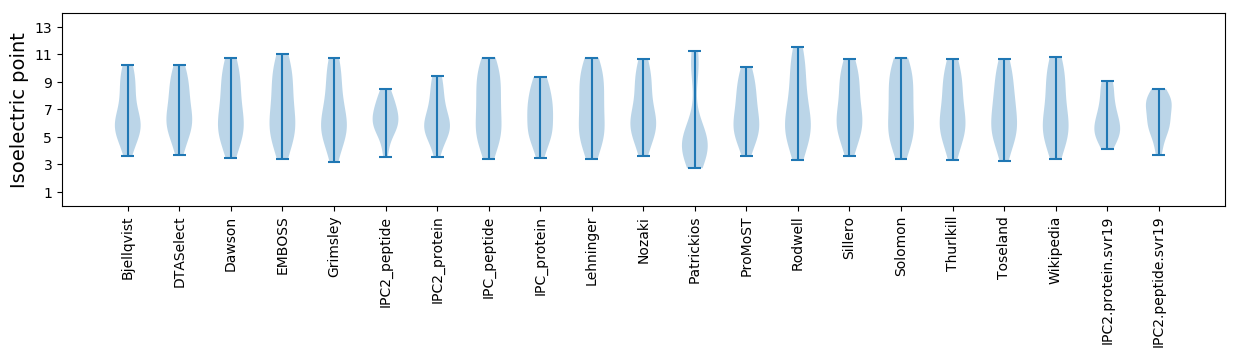
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZTC6|A0A4D5ZTC6_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan631 OX=2558795 GN=JavanS631_0004 PE=4 SV=1
MM1 pKa = 7.92KK2 pKa = 9.25ITEE5 pKa = 4.18YY6 pKa = 10.59KK7 pKa = 9.86KK8 pKa = 10.3KK9 pKa = 10.54NGTIVYY15 pKa = 9.63RR16 pKa = 11.84SQVYY20 pKa = 10.49LGVDD24 pKa = 4.02VITGKK29 pKa = 8.3EE30 pKa = 3.87VKK32 pKa = 9.42TRR34 pKa = 11.84ITARR38 pKa = 11.84TKK40 pKa = 10.91KK41 pKa = 9.72EE42 pKa = 3.87LKK44 pKa = 10.53LLAKK48 pKa = 10.07QKK50 pKa = 10.28QNEE53 pKa = 4.58FIRR56 pKa = 11.84NGSTVHH62 pKa = 7.16SEE64 pKa = 4.1IKK66 pKa = 9.79IKK68 pKa = 10.42SYY70 pKa = 11.4KK71 pKa = 10.18EE72 pKa = 3.81LTDD75 pKa = 3.74LWWNSYY81 pKa = 10.64KK82 pKa = 9.94NTVKK86 pKa = 10.5PNTVGSVSLLLKK98 pKa = 10.27NHH100 pKa = 7.51IIPTFGDD107 pKa = 3.54YY108 pKa = 10.98KK109 pKa = 10.46LDD111 pKa = 4.51KK112 pKa = 9.87ITTPLIQNKK121 pKa = 6.88VNKK124 pKa = 9.08WADD127 pKa = 3.24KK128 pKa = 11.0ANNNEE133 pKa = 4.02SGAFAHH139 pKa = 6.33YY140 pKa = 10.33DD141 pKa = 3.32KK142 pKa = 11.42LHH144 pKa = 6.45ALNKK148 pKa = 10.21RR149 pKa = 11.84ILQYY153 pKa = 11.04GVTMQLLDD161 pKa = 4.05NNPALNVVLPRR172 pKa = 11.84KK173 pKa = 9.6KK174 pKa = 10.18KK175 pKa = 10.15RR176 pKa = 11.84EE177 pKa = 4.04KK178 pKa = 10.17TSLKK182 pKa = 10.63YY183 pKa = 10.52FDD185 pKa = 3.4NTEE188 pKa = 3.94LKK190 pKa = 10.74KK191 pKa = 10.89FLDD194 pKa = 3.89YY195 pKa = 11.39LDD197 pKa = 4.99GLNQSNYY204 pKa = 7.31RR205 pKa = 11.84HH206 pKa = 6.04HH207 pKa = 7.45FEE209 pKa = 3.71VTLYY213 pKa = 10.92KK214 pKa = 10.49FLLATGCRR222 pKa = 11.84INEE225 pKa = 4.0ALALSWSDD233 pKa = 2.88IDD235 pKa = 5.27LEE237 pKa = 4.38SSSLSITKK245 pKa = 8.8TLNRR249 pKa = 11.84YY250 pKa = 9.35VEE252 pKa = 4.31VNTPKK257 pKa = 10.3SQASIRR263 pKa = 11.84TIDD266 pKa = 3.35IDD268 pKa = 3.57KK269 pKa = 9.93ATVLMLKK276 pKa = 9.72QYY278 pKa = 10.89RR279 pKa = 11.84NRR281 pKa = 11.84QRR283 pKa = 11.84IQGLEE288 pKa = 3.66IGLVPDD294 pKa = 3.87IVFSDD299 pKa = 4.82FINKK303 pKa = 8.57YY304 pKa = 11.04VNDD307 pKa = 3.4QTLYY311 pKa = 9.64TRR313 pKa = 11.84LRR315 pKa = 11.84THH317 pKa = 7.03FKK319 pKa = 10.33RR320 pKa = 11.84AGVSNIGFHH329 pKa = 6.37GFRR332 pKa = 11.84HH333 pKa = 4.66THH335 pKa = 5.52ATLLYY340 pKa = 10.37NAGIDD345 pKa = 3.78PKK347 pKa = 10.95ALQHH351 pKa = 6.52RR352 pKa = 11.84LGHH355 pKa = 5.34STISMTLDD363 pKa = 3.4TYY365 pKa = 11.55SHH367 pKa = 7.1LSKK370 pKa = 10.96EE371 pKa = 4.3NAKK374 pKa = 10.34KK375 pKa = 10.33AVSFFEE381 pKa = 4.35TAVSSLL387 pKa = 3.45
MM1 pKa = 7.92KK2 pKa = 9.25ITEE5 pKa = 4.18YY6 pKa = 10.59KK7 pKa = 9.86KK8 pKa = 10.3KK9 pKa = 10.54NGTIVYY15 pKa = 9.63RR16 pKa = 11.84SQVYY20 pKa = 10.49LGVDD24 pKa = 4.02VITGKK29 pKa = 8.3EE30 pKa = 3.87VKK32 pKa = 9.42TRR34 pKa = 11.84ITARR38 pKa = 11.84TKK40 pKa = 10.91KK41 pKa = 9.72EE42 pKa = 3.87LKK44 pKa = 10.53LLAKK48 pKa = 10.07QKK50 pKa = 10.28QNEE53 pKa = 4.58FIRR56 pKa = 11.84NGSTVHH62 pKa = 7.16SEE64 pKa = 4.1IKK66 pKa = 9.79IKK68 pKa = 10.42SYY70 pKa = 11.4KK71 pKa = 10.18EE72 pKa = 3.81LTDD75 pKa = 3.74LWWNSYY81 pKa = 10.64KK82 pKa = 9.94NTVKK86 pKa = 10.5PNTVGSVSLLLKK98 pKa = 10.27NHH100 pKa = 7.51IIPTFGDD107 pKa = 3.54YY108 pKa = 10.98KK109 pKa = 10.46LDD111 pKa = 4.51KK112 pKa = 9.87ITTPLIQNKK121 pKa = 6.88VNKK124 pKa = 9.08WADD127 pKa = 3.24KK128 pKa = 11.0ANNNEE133 pKa = 4.02SGAFAHH139 pKa = 6.33YY140 pKa = 10.33DD141 pKa = 3.32KK142 pKa = 11.42LHH144 pKa = 6.45ALNKK148 pKa = 10.21RR149 pKa = 11.84ILQYY153 pKa = 11.04GVTMQLLDD161 pKa = 4.05NNPALNVVLPRR172 pKa = 11.84KK173 pKa = 9.6KK174 pKa = 10.18KK175 pKa = 10.15RR176 pKa = 11.84EE177 pKa = 4.04KK178 pKa = 10.17TSLKK182 pKa = 10.63YY183 pKa = 10.52FDD185 pKa = 3.4NTEE188 pKa = 3.94LKK190 pKa = 10.74KK191 pKa = 10.89FLDD194 pKa = 3.89YY195 pKa = 11.39LDD197 pKa = 4.99GLNQSNYY204 pKa = 7.31RR205 pKa = 11.84HH206 pKa = 6.04HH207 pKa = 7.45FEE209 pKa = 3.71VTLYY213 pKa = 10.92KK214 pKa = 10.49FLLATGCRR222 pKa = 11.84INEE225 pKa = 4.0ALALSWSDD233 pKa = 2.88IDD235 pKa = 5.27LEE237 pKa = 4.38SSSLSITKK245 pKa = 8.8TLNRR249 pKa = 11.84YY250 pKa = 9.35VEE252 pKa = 4.31VNTPKK257 pKa = 10.3SQASIRR263 pKa = 11.84TIDD266 pKa = 3.35IDD268 pKa = 3.57KK269 pKa = 9.93ATVLMLKK276 pKa = 9.72QYY278 pKa = 10.89RR279 pKa = 11.84NRR281 pKa = 11.84QRR283 pKa = 11.84IQGLEE288 pKa = 3.66IGLVPDD294 pKa = 3.87IVFSDD299 pKa = 4.82FINKK303 pKa = 8.57YY304 pKa = 11.04VNDD307 pKa = 3.4QTLYY311 pKa = 9.64TRR313 pKa = 11.84LRR315 pKa = 11.84THH317 pKa = 7.03FKK319 pKa = 10.33RR320 pKa = 11.84AGVSNIGFHH329 pKa = 6.37GFRR332 pKa = 11.84HH333 pKa = 4.66THH335 pKa = 5.52ATLLYY340 pKa = 10.37NAGIDD345 pKa = 3.78PKK347 pKa = 10.95ALQHH351 pKa = 6.52RR352 pKa = 11.84LGHH355 pKa = 5.34STISMTLDD363 pKa = 3.4TYY365 pKa = 11.55SHH367 pKa = 7.1LSKK370 pKa = 10.96EE371 pKa = 4.3NAKK374 pKa = 10.34KK375 pKa = 10.33AVSFFEE381 pKa = 4.35TAVSSLL387 pKa = 3.45
Molecular weight: 44.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3518 |
48 |
672 |
195.4 |
22.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.827 ± 0.886 | 0.597 ± 0.213 |
6.083 ± 0.628 | 7.504 ± 0.864 |
4.434 ± 0.395 | 5.372 ± 1.057 |
1.45 ± 0.299 | 7.675 ± 0.323 |
9.608 ± 0.899 | 10.432 ± 0.476 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.757 ± 0.341 | 6.737 ± 0.361 |
2.16 ± 0.196 | 3.894 ± 0.406 |
3.61 ± 0.53 | 6.652 ± 0.703 |
5.259 ± 0.443 | 5.429 ± 0.242 |
0.711 ± 0.104 | 3.809 ± 0.53 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
