
Sphingobium amiense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingobium
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
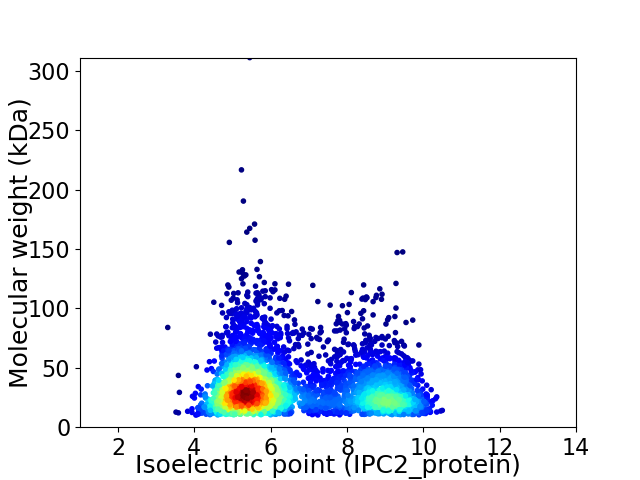
Virtual 2D-PAGE plot for 3954 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
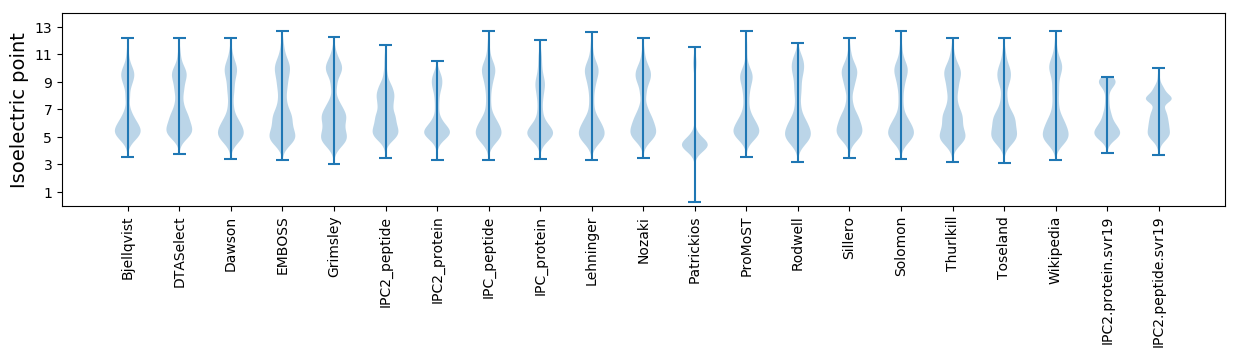
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A494W7Y4|A0A494W7Y4_9SPHN DUF3617 domain-containing protein OS=Sphingobium amiense OX=135719 GN=SAMIE_1027590 PE=4 SV=1
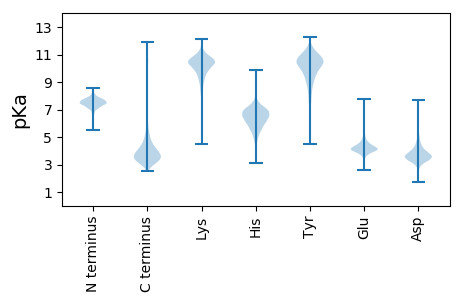
MM1 pKa = 7.79PKK3 pKa = 10.4LIVVNRR9 pKa = 11.84AGEE12 pKa = 3.97EE13 pKa = 3.86QEE15 pKa = 4.08IDD17 pKa = 3.64GEE19 pKa = 4.58TGLSVMEE26 pKa = 4.73VIRR29 pKa = 11.84DD30 pKa = 3.56HH31 pKa = 7.32GFDD34 pKa = 4.23EE35 pKa = 5.43LLALCGGCCSCATCHH50 pKa = 6.48VYY52 pKa = 10.52VDD54 pKa = 3.85PAFAGALPPMSEE66 pKa = 5.07DD67 pKa = 3.85EE68 pKa = 4.43NDD70 pKa = 4.07LLDD73 pKa = 4.81SSDD76 pKa = 3.59HH77 pKa = 6.39RR78 pKa = 11.84NEE80 pKa = 3.85TSRR83 pKa = 11.84LSCQVQLTGALDD95 pKa = 3.67GLRR98 pKa = 11.84VTIAPEE104 pKa = 3.78DD105 pKa = 3.45
MM1 pKa = 7.79PKK3 pKa = 10.4LIVVNRR9 pKa = 11.84AGEE12 pKa = 3.97EE13 pKa = 3.86QEE15 pKa = 4.08IDD17 pKa = 3.64GEE19 pKa = 4.58TGLSVMEE26 pKa = 4.73VIRR29 pKa = 11.84DD30 pKa = 3.56HH31 pKa = 7.32GFDD34 pKa = 4.23EE35 pKa = 5.43LLALCGGCCSCATCHH50 pKa = 6.48VYY52 pKa = 10.52VDD54 pKa = 3.85PAFAGALPPMSEE66 pKa = 5.07DD67 pKa = 3.85EE68 pKa = 4.43NDD70 pKa = 4.07LLDD73 pKa = 4.81SSDD76 pKa = 3.59HH77 pKa = 6.39RR78 pKa = 11.84NEE80 pKa = 3.85TSRR83 pKa = 11.84LSCQVQLTGALDD95 pKa = 3.67GLRR98 pKa = 11.84VTIAPEE104 pKa = 3.78DD105 pKa = 3.45
Molecular weight: 11.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A494WAD8|A0A494WAD8_9SPHN WYL domain-containing protein OS=Sphingobium amiense OX=135719 GN=SAMIE_1031180 PE=4 SV=1
MM1 pKa = 7.07TALATHH7 pKa = 7.09LAAFLRR13 pKa = 11.84EE14 pKa = 3.96HH15 pKa = 7.16LPRR18 pKa = 11.84EE19 pKa = 4.51RR20 pKa = 11.84NVSPHH25 pKa = 5.41TVTTYY30 pKa = 11.44ANCFALLVRR39 pKa = 11.84FAANHH44 pKa = 6.14LKK46 pKa = 10.3RR47 pKa = 11.84RR48 pKa = 11.84PTDD51 pKa = 3.35LTIEE55 pKa = 4.55DD56 pKa = 4.72FDD58 pKa = 4.7PALIMAFLDD67 pKa = 3.46HH68 pKa = 7.92SEE70 pKa = 4.19EE71 pKa = 3.93ARR73 pKa = 11.84KK74 pKa = 10.01NSPRR78 pKa = 11.84TRR80 pKa = 11.84NARR83 pKa = 11.84LAAIRR88 pKa = 11.84AFFRR92 pKa = 11.84YY93 pKa = 9.38VEE95 pKa = 4.05YY96 pKa = 10.4RR97 pKa = 11.84VPACLDD103 pKa = 3.15LALRR107 pKa = 11.84VRR109 pKa = 11.84AVPTKK114 pKa = 10.23RR115 pKa = 11.84TDD117 pKa = 3.1TTLIDD122 pKa = 3.61YY123 pKa = 7.56LTRR126 pKa = 11.84DD127 pKa = 4.29EE128 pKa = 4.47ITALLDD134 pKa = 3.88APDD137 pKa = 4.11PQSRR141 pKa = 11.84LGTRR145 pKa = 11.84DD146 pKa = 2.87RR147 pKa = 11.84AMLHH151 pKa = 6.15LAYY154 pKa = 10.37AGGLRR159 pKa = 11.84VAEE162 pKa = 4.2LLSLQMQDD170 pKa = 3.17FPEE173 pKa = 4.79RR174 pKa = 11.84SLATVHH180 pKa = 6.97IIGKK184 pKa = 8.23GRR186 pKa = 11.84RR187 pKa = 11.84EE188 pKa = 4.05RR189 pKa = 11.84ILPLWRR195 pKa = 11.84EE196 pKa = 4.12TQASLRR202 pKa = 11.84AWLAVRR208 pKa = 11.84PQCQAMEE215 pKa = 4.28IFLNAHH221 pKa = 6.44GEE223 pKa = 3.96PMTRR227 pKa = 11.84DD228 pKa = 3.04GFAFRR233 pKa = 11.84LAKK236 pKa = 10.24HH237 pKa = 5.52VATAAKK243 pKa = 9.25KK244 pKa = 10.13QPSLLRR250 pKa = 11.84KK251 pKa = 9.25RR252 pKa = 11.84VTPHH256 pKa = 5.67VLRR259 pKa = 11.84HH260 pKa = 4.92SCAMHH265 pKa = 6.13TLAATGDD272 pKa = 3.51IRR274 pKa = 11.84KK275 pKa = 8.74VALWLGHH282 pKa = 6.83ASIQSTEE289 pKa = 3.76AYY291 pKa = 10.05LRR293 pKa = 11.84ADD295 pKa = 3.93PEE297 pKa = 4.18EE298 pKa = 4.21KK299 pKa = 10.66LQILAAHH306 pKa = 7.17GSPLVKK312 pKa = 9.78PGRR315 pKa = 11.84FRR317 pKa = 11.84PPADD321 pKa = 3.92PLIMMLNDD329 pKa = 2.95VRR331 pKa = 11.84KK332 pKa = 9.13RR333 pKa = 11.84ASPRR337 pKa = 11.84GRR339 pKa = 11.84SAPGIQSPP347 pKa = 3.87
MM1 pKa = 7.07TALATHH7 pKa = 7.09LAAFLRR13 pKa = 11.84EE14 pKa = 3.96HH15 pKa = 7.16LPRR18 pKa = 11.84EE19 pKa = 4.51RR20 pKa = 11.84NVSPHH25 pKa = 5.41TVTTYY30 pKa = 11.44ANCFALLVRR39 pKa = 11.84FAANHH44 pKa = 6.14LKK46 pKa = 10.3RR47 pKa = 11.84RR48 pKa = 11.84PTDD51 pKa = 3.35LTIEE55 pKa = 4.55DD56 pKa = 4.72FDD58 pKa = 4.7PALIMAFLDD67 pKa = 3.46HH68 pKa = 7.92SEE70 pKa = 4.19EE71 pKa = 3.93ARR73 pKa = 11.84KK74 pKa = 10.01NSPRR78 pKa = 11.84TRR80 pKa = 11.84NARR83 pKa = 11.84LAAIRR88 pKa = 11.84AFFRR92 pKa = 11.84YY93 pKa = 9.38VEE95 pKa = 4.05YY96 pKa = 10.4RR97 pKa = 11.84VPACLDD103 pKa = 3.15LALRR107 pKa = 11.84VRR109 pKa = 11.84AVPTKK114 pKa = 10.23RR115 pKa = 11.84TDD117 pKa = 3.1TTLIDD122 pKa = 3.61YY123 pKa = 7.56LTRR126 pKa = 11.84DD127 pKa = 4.29EE128 pKa = 4.47ITALLDD134 pKa = 3.88APDD137 pKa = 4.11PQSRR141 pKa = 11.84LGTRR145 pKa = 11.84DD146 pKa = 2.87RR147 pKa = 11.84AMLHH151 pKa = 6.15LAYY154 pKa = 10.37AGGLRR159 pKa = 11.84VAEE162 pKa = 4.2LLSLQMQDD170 pKa = 3.17FPEE173 pKa = 4.79RR174 pKa = 11.84SLATVHH180 pKa = 6.97IIGKK184 pKa = 8.23GRR186 pKa = 11.84RR187 pKa = 11.84EE188 pKa = 4.05RR189 pKa = 11.84ILPLWRR195 pKa = 11.84EE196 pKa = 4.12TQASLRR202 pKa = 11.84AWLAVRR208 pKa = 11.84PQCQAMEE215 pKa = 4.28IFLNAHH221 pKa = 6.44GEE223 pKa = 3.96PMTRR227 pKa = 11.84DD228 pKa = 3.04GFAFRR233 pKa = 11.84LAKK236 pKa = 10.24HH237 pKa = 5.52VATAAKK243 pKa = 9.25KK244 pKa = 10.13QPSLLRR250 pKa = 11.84KK251 pKa = 9.25RR252 pKa = 11.84VTPHH256 pKa = 5.67VLRR259 pKa = 11.84HH260 pKa = 4.92SCAMHH265 pKa = 6.13TLAATGDD272 pKa = 3.51IRR274 pKa = 11.84KK275 pKa = 8.74VALWLGHH282 pKa = 6.83ASIQSTEE289 pKa = 3.76AYY291 pKa = 10.05LRR293 pKa = 11.84ADD295 pKa = 3.93PEE297 pKa = 4.18EE298 pKa = 4.21KK299 pKa = 10.66LQILAAHH306 pKa = 7.17GSPLVKK312 pKa = 9.78PGRR315 pKa = 11.84FRR317 pKa = 11.84PPADD321 pKa = 3.92PLIMMLNDD329 pKa = 2.95VRR331 pKa = 11.84KK332 pKa = 9.13RR333 pKa = 11.84ASPRR337 pKa = 11.84GRR339 pKa = 11.84SAPGIQSPP347 pKa = 3.87
Molecular weight: 39.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1349417 |
100 |
2803 |
341.3 |
36.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.277 ± 0.063 | 0.795 ± 0.012 |
6.055 ± 0.029 | 5.302 ± 0.035 |
3.512 ± 0.026 | 8.79 ± 0.039 |
2.078 ± 0.02 | 5.013 ± 0.025 |
2.896 ± 0.028 | 9.979 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.511 ± 0.019 | 2.455 ± 0.024 |
5.372 ± 0.03 | 3.224 ± 0.021 |
7.622 ± 0.043 | 5.304 ± 0.027 |
5.078 ± 0.028 | 7.024 ± 0.031 |
1.45 ± 0.015 | 2.262 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
