
Beutenbergia cavernae (strain ATCC BAA-8 / DSM 12333 / NBRC 16432)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Beutenbergiaceae; Beutenbergia; Beutenbergia cavernae
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
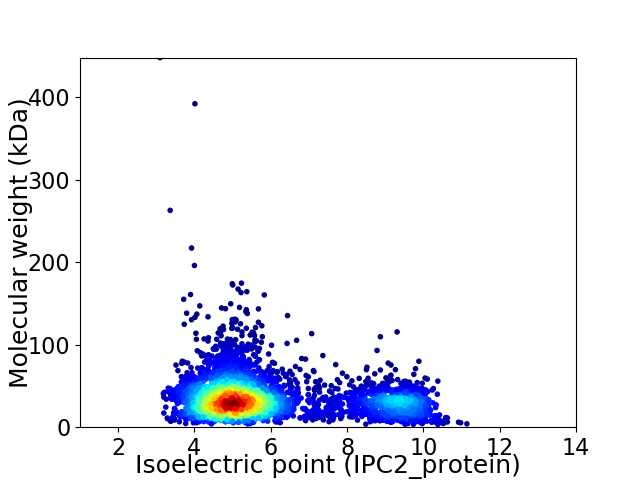
Virtual 2D-PAGE plot for 4195 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C5BY60|C5BY60_BEUC1 Major facilitator superfamily MFS_1 OS=Beutenbergia cavernae (strain ATCC BAA-8 / DSM 12333 / NBRC 16432) OX=471853 GN=Bcav_2715 PE=4 SV=1
MM1 pKa = 7.01SRR3 pKa = 11.84AWRR6 pKa = 11.84EE7 pKa = 3.66GPYY10 pKa = 10.33ADD12 pKa = 3.22PVSRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84AARR21 pKa = 11.84GVAAVLAVAGLAAACAGPAPMLQPVDD47 pKa = 4.06AGAPTDD53 pKa = 4.07PGQQGSATPQPSDD66 pKa = 3.28DD67 pKa = 4.29ALAEE71 pKa = 4.61FYY73 pKa = 10.08TQVPSWYY80 pKa = 8.58ACGGGFEE87 pKa = 4.45CADD90 pKa = 3.36VEE92 pKa = 5.1VPLDD96 pKa = 3.49YY97 pKa = 10.83DD98 pKa = 3.62APDD101 pKa = 3.77GEE103 pKa = 4.43RR104 pKa = 11.84VQLAVKK110 pKa = 10.22RR111 pKa = 11.84LPADD115 pKa = 3.8DD116 pKa = 3.78PAARR120 pKa = 11.84QGSLLVNPGGPGASGLDD137 pKa = 3.9LVDD140 pKa = 3.71SAADD144 pKa = 3.55LFSNRR149 pKa = 11.84VLDD152 pKa = 4.52AYY154 pKa = 10.69DD155 pKa = 3.35VVGFDD160 pKa = 3.64PRR162 pKa = 11.84GVGSSTKK169 pKa = 9.7IVCLGPDD176 pKa = 3.53EE177 pKa = 4.9EE178 pKa = 5.27DD179 pKa = 3.86PSGGDD184 pKa = 3.35YY185 pKa = 11.14DD186 pKa = 5.42LSDD189 pKa = 3.91DD190 pKa = 5.04AEE192 pKa = 4.36VQRR195 pKa = 11.84LVDD198 pKa = 4.89DD199 pKa = 4.93LDD201 pKa = 4.77ALGEE205 pKa = 4.07MCRR208 pKa = 11.84DD209 pKa = 3.41HH210 pKa = 8.93SGDD213 pKa = 4.11LLDD216 pKa = 5.56HH217 pKa = 6.9VDD219 pKa = 3.74TVHH222 pKa = 7.72AARR225 pKa = 11.84DD226 pKa = 3.76LDD228 pKa = 3.72VLRR231 pKa = 11.84AVLGDD236 pKa = 3.48EE237 pKa = 3.9RR238 pKa = 11.84LTYY241 pKa = 10.81LGFSYY246 pKa = 9.06GTILGATFAEE256 pKa = 4.62LFPDD260 pKa = 3.32RR261 pKa = 11.84VGRR264 pKa = 11.84LVLDD268 pKa = 4.13GAIDD272 pKa = 3.75PSIDD276 pKa = 3.66YY277 pKa = 8.45TQMTADD283 pKa = 3.2QVDD286 pKa = 3.88GFEE289 pKa = 4.24VAFRR293 pKa = 11.84SYY295 pKa = 10.91LANCLEE301 pKa = 4.5GDD303 pKa = 3.61ACPFSGTEE311 pKa = 3.72DD312 pKa = 3.35EE313 pKa = 4.97AYY315 pKa = 10.63DD316 pKa = 3.54RR317 pKa = 11.84AVAFLEE323 pKa = 4.18EE324 pKa = 4.91LDD326 pKa = 4.72AEE328 pKa = 4.39PLPSEE333 pKa = 4.71GEE335 pKa = 4.15DD336 pKa = 3.93GEE338 pKa = 4.5LTSDD342 pKa = 3.46EE343 pKa = 4.76AYY345 pKa = 10.52GAIQSSMYY353 pKa = 9.85VAWAWEE359 pKa = 3.81ALSEE363 pKa = 4.33GFTQAFEE370 pKa = 4.53DD371 pKa = 4.21ADD373 pKa = 4.14GSALDD378 pKa = 4.52VIAHH382 pKa = 6.42EE383 pKa = 5.18NDD385 pKa = 4.03DD386 pKa = 4.62PDD388 pKa = 4.47PNADD392 pKa = 3.37FAFWGIDD399 pKa = 3.29CSDD402 pKa = 3.72YY403 pKa = 11.04PITSSVDD410 pKa = 3.5EE411 pKa = 4.71ILAQADD417 pKa = 3.85EE418 pKa = 4.73LEE420 pKa = 4.34EE421 pKa = 4.64ASALFGAAMGTGEE434 pKa = 4.5LVCRR438 pKa = 11.84QWPYY442 pKa = 10.08QSTAVRR448 pKa = 11.84EE449 pKa = 4.32PIEE452 pKa = 4.24AAGAAPILVVGTTRR466 pKa = 11.84DD467 pKa = 3.1PATPYY472 pKa = 10.25RR473 pKa = 11.84WAEE476 pKa = 3.74ALAGQLEE483 pKa = 4.44SGRR486 pKa = 11.84LLTWDD491 pKa = 3.76GDD493 pKa = 3.57GHH495 pKa = 5.17TAYY498 pKa = 10.44ASGSPCIDD506 pKa = 3.56EE507 pKa = 5.37AVQAYY512 pKa = 10.24LLDD515 pKa = 3.84GTLPDD520 pKa = 5.51DD521 pKa = 4.14GTVCC525 pKa = 4.7
MM1 pKa = 7.01SRR3 pKa = 11.84AWRR6 pKa = 11.84EE7 pKa = 3.66GPYY10 pKa = 10.33ADD12 pKa = 3.22PVSRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84AARR21 pKa = 11.84GVAAVLAVAGLAAACAGPAPMLQPVDD47 pKa = 4.06AGAPTDD53 pKa = 4.07PGQQGSATPQPSDD66 pKa = 3.28DD67 pKa = 4.29ALAEE71 pKa = 4.61FYY73 pKa = 10.08TQVPSWYY80 pKa = 8.58ACGGGFEE87 pKa = 4.45CADD90 pKa = 3.36VEE92 pKa = 5.1VPLDD96 pKa = 3.49YY97 pKa = 10.83DD98 pKa = 3.62APDD101 pKa = 3.77GEE103 pKa = 4.43RR104 pKa = 11.84VQLAVKK110 pKa = 10.22RR111 pKa = 11.84LPADD115 pKa = 3.8DD116 pKa = 3.78PAARR120 pKa = 11.84QGSLLVNPGGPGASGLDD137 pKa = 3.9LVDD140 pKa = 3.71SAADD144 pKa = 3.55LFSNRR149 pKa = 11.84VLDD152 pKa = 4.52AYY154 pKa = 10.69DD155 pKa = 3.35VVGFDD160 pKa = 3.64PRR162 pKa = 11.84GVGSSTKK169 pKa = 9.7IVCLGPDD176 pKa = 3.53EE177 pKa = 4.9EE178 pKa = 5.27DD179 pKa = 3.86PSGGDD184 pKa = 3.35YY185 pKa = 11.14DD186 pKa = 5.42LSDD189 pKa = 3.91DD190 pKa = 5.04AEE192 pKa = 4.36VQRR195 pKa = 11.84LVDD198 pKa = 4.89DD199 pKa = 4.93LDD201 pKa = 4.77ALGEE205 pKa = 4.07MCRR208 pKa = 11.84DD209 pKa = 3.41HH210 pKa = 8.93SGDD213 pKa = 4.11LLDD216 pKa = 5.56HH217 pKa = 6.9VDD219 pKa = 3.74TVHH222 pKa = 7.72AARR225 pKa = 11.84DD226 pKa = 3.76LDD228 pKa = 3.72VLRR231 pKa = 11.84AVLGDD236 pKa = 3.48EE237 pKa = 3.9RR238 pKa = 11.84LTYY241 pKa = 10.81LGFSYY246 pKa = 9.06GTILGATFAEE256 pKa = 4.62LFPDD260 pKa = 3.32RR261 pKa = 11.84VGRR264 pKa = 11.84LVLDD268 pKa = 4.13GAIDD272 pKa = 3.75PSIDD276 pKa = 3.66YY277 pKa = 8.45TQMTADD283 pKa = 3.2QVDD286 pKa = 3.88GFEE289 pKa = 4.24VAFRR293 pKa = 11.84SYY295 pKa = 10.91LANCLEE301 pKa = 4.5GDD303 pKa = 3.61ACPFSGTEE311 pKa = 3.72DD312 pKa = 3.35EE313 pKa = 4.97AYY315 pKa = 10.63DD316 pKa = 3.54RR317 pKa = 11.84AVAFLEE323 pKa = 4.18EE324 pKa = 4.91LDD326 pKa = 4.72AEE328 pKa = 4.39PLPSEE333 pKa = 4.71GEE335 pKa = 4.15DD336 pKa = 3.93GEE338 pKa = 4.5LTSDD342 pKa = 3.46EE343 pKa = 4.76AYY345 pKa = 10.52GAIQSSMYY353 pKa = 9.85VAWAWEE359 pKa = 3.81ALSEE363 pKa = 4.33GFTQAFEE370 pKa = 4.53DD371 pKa = 4.21ADD373 pKa = 4.14GSALDD378 pKa = 4.52VIAHH382 pKa = 6.42EE383 pKa = 5.18NDD385 pKa = 4.03DD386 pKa = 4.62PDD388 pKa = 4.47PNADD392 pKa = 3.37FAFWGIDD399 pKa = 3.29CSDD402 pKa = 3.72YY403 pKa = 11.04PITSSVDD410 pKa = 3.5EE411 pKa = 4.71ILAQADD417 pKa = 3.85EE418 pKa = 4.73LEE420 pKa = 4.34EE421 pKa = 4.64ASALFGAAMGTGEE434 pKa = 4.5LVCRR438 pKa = 11.84QWPYY442 pKa = 10.08QSTAVRR448 pKa = 11.84EE449 pKa = 4.32PIEE452 pKa = 4.24AAGAAPILVVGTTRR466 pKa = 11.84DD467 pKa = 3.1PATPYY472 pKa = 10.25RR473 pKa = 11.84WAEE476 pKa = 3.74ALAGQLEE483 pKa = 4.44SGRR486 pKa = 11.84LLTWDD491 pKa = 3.76GDD493 pKa = 3.57GHH495 pKa = 5.17TAYY498 pKa = 10.44ASGSPCIDD506 pKa = 3.56EE507 pKa = 5.37AVQAYY512 pKa = 10.24LLDD515 pKa = 3.84GTLPDD520 pKa = 5.51DD521 pKa = 4.14GTVCC525 pKa = 4.7
Molecular weight: 55.44 kDa
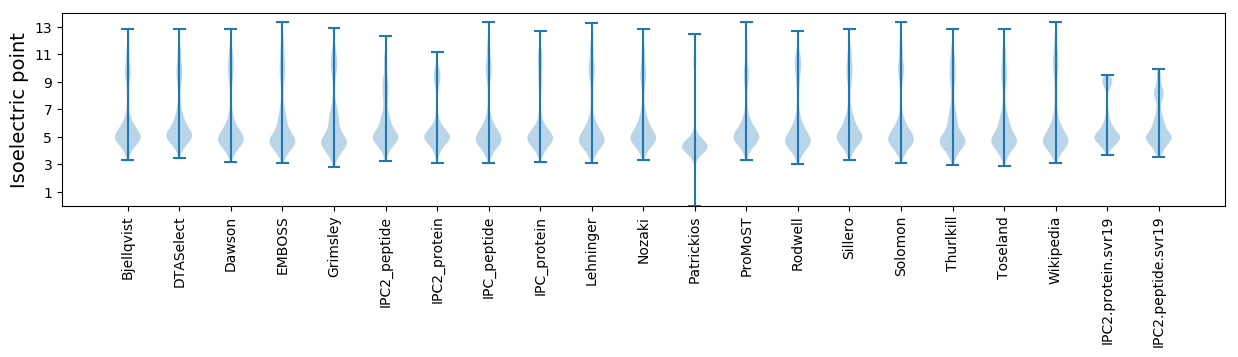
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C5C197|C5C197_BEUC1 Thiamine_BP domain-containing protein OS=Beutenbergia cavernae (strain ATCC BAA-8 / DSM 12333 / NBRC 16432) OX=471853 GN=Bcav_3264 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1434007 |
32 |
4672 |
341.8 |
36.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.644 ± 0.058 | 0.52 ± 0.008 |
6.634 ± 0.036 | 5.488 ± 0.036 |
2.761 ± 0.024 | 9.442 ± 0.037 |
2.031 ± 0.021 | 3.216 ± 0.025 |
1.088 ± 0.02 | 10.244 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.597 ± 0.014 | 1.507 ± 0.017 |
5.999 ± 0.031 | 2.403 ± 0.02 |
7.836 ± 0.047 | 5.057 ± 0.024 |
6.131 ± 0.057 | 9.814 ± 0.037 |
1.655 ± 0.019 | 1.932 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
