
Fruit bat alphaherpesvirus 1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Simplexvirus; Pteropodid alphaherpesvirus 1
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
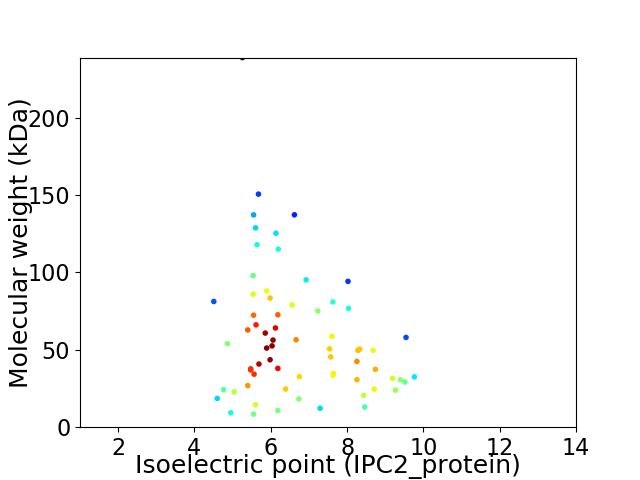
Virtual 2D-PAGE plot for 68 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A060Q0V3|A0A060Q0V3_9ALPH Capsid vertex component 2 OS=Fruit bat alphaherpesvirus 1 OX=1343901 GN=UL25 PE=3 SV=1
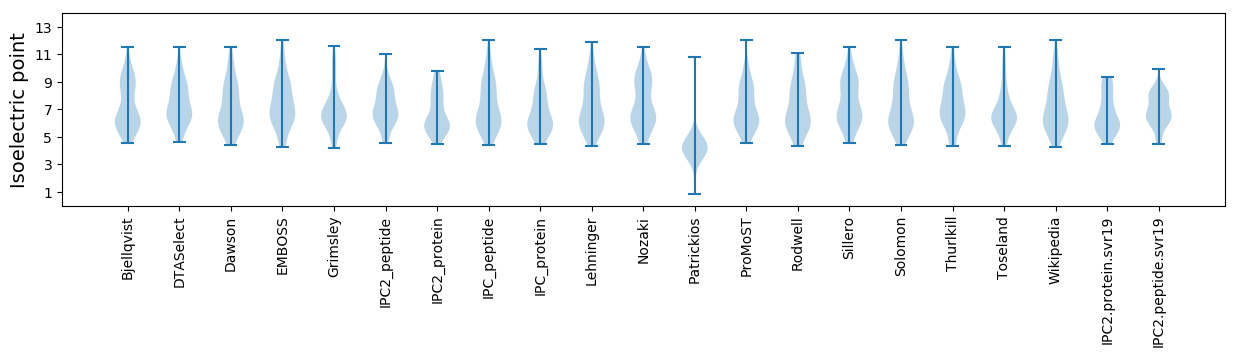
MM1 pKa = 7.98RR2 pKa = 11.84DD3 pKa = 4.2DD4 pKa = 5.58LPLIDD9 pKa = 5.22PLLVDD14 pKa = 3.79EE15 pKa = 5.3TEE17 pKa = 4.29SSEE20 pKa = 5.37DD21 pKa = 3.75PDD23 pKa = 3.98DD24 pKa = 5.58LSIEE28 pKa = 4.29EE29 pKa = 4.12QFSLSSYY36 pKa = 8.1GTADD40 pKa = 3.61FFVSSAYY47 pKa = 10.45SRR49 pKa = 11.84LPSHH53 pKa = 6.23TQPVFSKK60 pKa = 10.24RR61 pKa = 11.84VFVFVLAFFFLKK73 pKa = 10.35PFQLASLCVYY83 pKa = 9.23YY84 pKa = 10.94HH85 pKa = 4.83EE86 pKa = 4.7TGRR89 pKa = 11.84LAALACVAAAVLGYY103 pKa = 10.25YY104 pKa = 8.34VLWCARR110 pKa = 11.84ACALYY115 pKa = 10.2WNIKK119 pKa = 7.31QDD121 pKa = 4.1RR122 pKa = 11.84LPLTPTAFWFLVVCFSGLALGALCAAAHH150 pKa = 5.62EE151 pKa = 4.53TFAVDD156 pKa = 4.59GLFDD160 pKa = 6.03LISTGQVLPPTDD172 pKa = 3.83PLRR175 pKa = 11.84VRR177 pKa = 11.84LLTIACVTMVGLWVAAADD195 pKa = 4.09SFAVAANFFLARR207 pKa = 11.84FWTRR211 pKa = 11.84AILNAPVAFF220 pKa = 4.58
MM1 pKa = 7.98RR2 pKa = 11.84DD3 pKa = 4.2DD4 pKa = 5.58LPLIDD9 pKa = 5.22PLLVDD14 pKa = 3.79EE15 pKa = 5.3TEE17 pKa = 4.29SSEE20 pKa = 5.37DD21 pKa = 3.75PDD23 pKa = 3.98DD24 pKa = 5.58LSIEE28 pKa = 4.29EE29 pKa = 4.12QFSLSSYY36 pKa = 8.1GTADD40 pKa = 3.61FFVSSAYY47 pKa = 10.45SRR49 pKa = 11.84LPSHH53 pKa = 6.23TQPVFSKK60 pKa = 10.24RR61 pKa = 11.84VFVFVLAFFFLKK73 pKa = 10.35PFQLASLCVYY83 pKa = 9.23YY84 pKa = 10.94HH85 pKa = 4.83EE86 pKa = 4.7TGRR89 pKa = 11.84LAALACVAAAVLGYY103 pKa = 10.25YY104 pKa = 8.34VLWCARR110 pKa = 11.84ACALYY115 pKa = 10.2WNIKK119 pKa = 7.31QDD121 pKa = 4.1RR122 pKa = 11.84LPLTPTAFWFLVVCFSGLALGALCAAAHH150 pKa = 5.62EE151 pKa = 4.53TFAVDD156 pKa = 4.59GLFDD160 pKa = 6.03LISTGQVLPPTDD172 pKa = 3.83PLRR175 pKa = 11.84VRR177 pKa = 11.84LLTIACVTMVGLWVAAADD195 pKa = 4.09SFAVAANFFLARR207 pKa = 11.84FWTRR211 pKa = 11.84AILNAPVAFF220 pKa = 4.58
Molecular weight: 24.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A060PYC0|A0A060PYC0_9ALPH Envelope glycoprotein K OS=Fruit bat alphaherpesvirus 1 OX=1343901 GN=UL53 PE=3 SV=1
MM1 pKa = 7.11ATRR4 pKa = 11.84RR5 pKa = 11.84TARR8 pKa = 11.84PPARR12 pKa = 11.84TPAPSRR18 pKa = 11.84PVLEE22 pKa = 5.24RR23 pKa = 11.84YY24 pKa = 9.48ASSSSTSTRR33 pKa = 11.84GGSIYY38 pKa = 10.32DD39 pKa = 3.42TDD41 pKa = 4.02DD42 pKa = 3.85RR43 pKa = 11.84PGGEE47 pKa = 4.09VQRR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 4.57YY53 pKa = 9.38HH54 pKa = 4.2QCEE57 pKa = 4.11EE58 pKa = 4.07DD59 pKa = 3.31WPVYY63 pKa = 10.38SDD65 pKa = 3.67SFEE68 pKa = 4.29SSSDD72 pKa = 3.88DD73 pKa = 3.68EE74 pKa = 5.75LDD76 pKa = 3.61AAMSAPTPSEE86 pKa = 3.48PTGRR90 pKa = 11.84GRR92 pKa = 11.84GAGRR96 pKa = 11.84SAAPRR101 pKa = 11.84APAARR106 pKa = 11.84RR107 pKa = 11.84APASGGQTNTRR118 pKa = 11.84GTRR121 pKa = 11.84GTAANPPTSRR131 pKa = 11.84PPALEE136 pKa = 4.26PDD138 pKa = 3.53ADD140 pKa = 3.71VTARR144 pKa = 11.84AGQGRR149 pKa = 11.84RR150 pKa = 11.84ARR152 pKa = 11.84KK153 pKa = 8.56PATKK157 pKa = 9.37PAPPPTIDD165 pKa = 3.53SQAKK169 pKa = 9.77GLTASNLLQFSGAPSSPTSPWNGRR193 pKa = 11.84TAIFNKK199 pKa = 9.23RR200 pKa = 11.84TFCAAVGRR208 pKa = 11.84VAAKK212 pKa = 9.92HH213 pKa = 5.54ARR215 pKa = 11.84QAAAKK220 pKa = 8.87LWDD223 pKa = 3.65MSNPQNDD230 pKa = 3.83EE231 pKa = 4.02EE232 pKa = 5.42LNTLLGATNIRR243 pKa = 11.84ITVCEE248 pKa = 4.16GQDD251 pKa = 3.63LLPRR255 pKa = 11.84ANDD258 pKa = 3.83LVTGACPEE266 pKa = 4.29PEE268 pKa = 4.34STSRR272 pKa = 11.84TPARR276 pKa = 11.84SVARR280 pKa = 11.84STSRR284 pKa = 11.84ARR286 pKa = 11.84RR287 pKa = 11.84PNN289 pKa = 3.15
MM1 pKa = 7.11ATRR4 pKa = 11.84RR5 pKa = 11.84TARR8 pKa = 11.84PPARR12 pKa = 11.84TPAPSRR18 pKa = 11.84PVLEE22 pKa = 5.24RR23 pKa = 11.84YY24 pKa = 9.48ASSSSTSTRR33 pKa = 11.84GGSIYY38 pKa = 10.32DD39 pKa = 3.42TDD41 pKa = 4.02DD42 pKa = 3.85RR43 pKa = 11.84PGGEE47 pKa = 4.09VQRR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 4.57YY53 pKa = 9.38HH54 pKa = 4.2QCEE57 pKa = 4.11EE58 pKa = 4.07DD59 pKa = 3.31WPVYY63 pKa = 10.38SDD65 pKa = 3.67SFEE68 pKa = 4.29SSSDD72 pKa = 3.88DD73 pKa = 3.68EE74 pKa = 5.75LDD76 pKa = 3.61AAMSAPTPSEE86 pKa = 3.48PTGRR90 pKa = 11.84GRR92 pKa = 11.84GAGRR96 pKa = 11.84SAAPRR101 pKa = 11.84APAARR106 pKa = 11.84RR107 pKa = 11.84APASGGQTNTRR118 pKa = 11.84GTRR121 pKa = 11.84GTAANPPTSRR131 pKa = 11.84PPALEE136 pKa = 4.26PDD138 pKa = 3.53ADD140 pKa = 3.71VTARR144 pKa = 11.84AGQGRR149 pKa = 11.84RR150 pKa = 11.84ARR152 pKa = 11.84KK153 pKa = 8.56PATKK157 pKa = 9.37PAPPPTIDD165 pKa = 3.53SQAKK169 pKa = 9.77GLTASNLLQFSGAPSSPTSPWNGRR193 pKa = 11.84TAIFNKK199 pKa = 9.23RR200 pKa = 11.84TFCAAVGRR208 pKa = 11.84VAAKK212 pKa = 9.92HH213 pKa = 5.54ARR215 pKa = 11.84QAAAKK220 pKa = 8.87LWDD223 pKa = 3.65MSNPQNDD230 pKa = 3.83EE231 pKa = 4.02EE232 pKa = 5.42LNTLLGATNIRR243 pKa = 11.84ITVCEE248 pKa = 4.16GQDD251 pKa = 3.63LLPRR255 pKa = 11.84ANDD258 pKa = 3.83LVTGACPEE266 pKa = 4.29PEE268 pKa = 4.34STSRR272 pKa = 11.84TPARR276 pKa = 11.84SVARR280 pKa = 11.84STSRR284 pKa = 11.84ARR286 pKa = 11.84RR287 pKa = 11.84PNN289 pKa = 3.15
Molecular weight: 30.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
36182 |
81 |
2219 |
532.1 |
58.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.321 ± 0.461 | 1.852 ± 0.13 |
4.679 ± 0.134 | 5.384 ± 0.189 |
3.958 ± 0.188 | 5.845 ± 0.18 |
2.653 ± 0.113 | 3.596 ± 0.205 |
2.36 ± 0.112 | 10.56 ± 0.318 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.755 ± 0.098 | 2.659 ± 0.157 |
8.018 ± 0.317 | 3.839 ± 0.126 |
7.258 ± 0.244 | 7.728 ± 0.277 |
6.362 ± 0.247 | 6.42 ± 0.169 |
1.205 ± 0.087 | 2.548 ± 0.142 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
