
Sewage-associated gemycircularvirus-10b
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus humas1; Human associated gemykibivirus 1
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
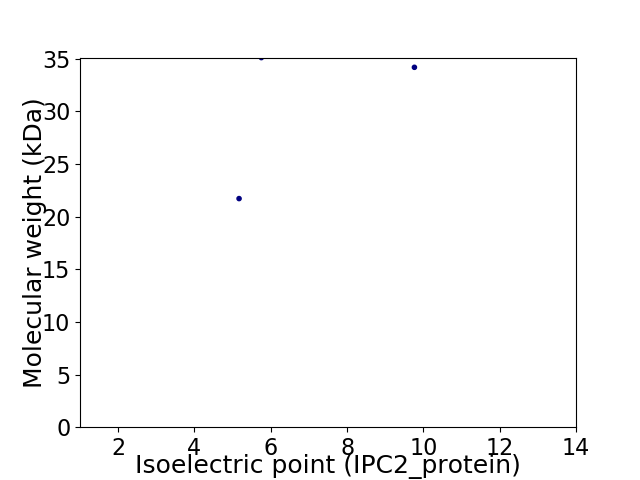
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
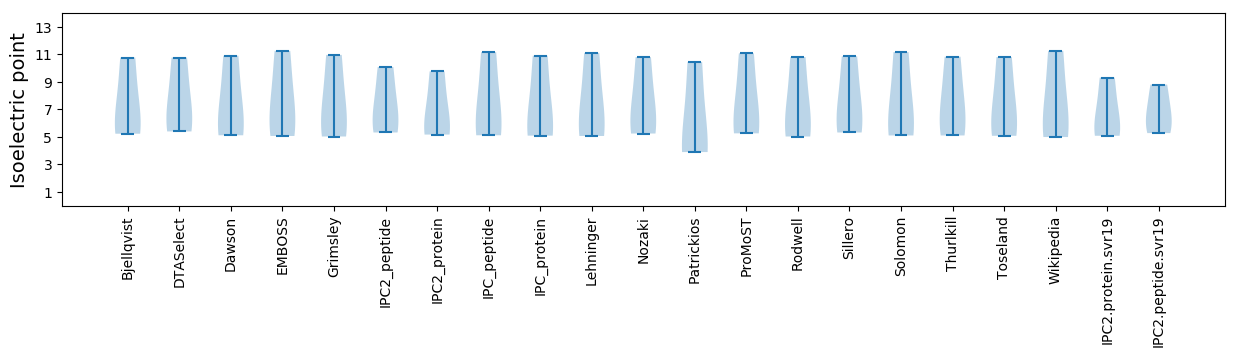
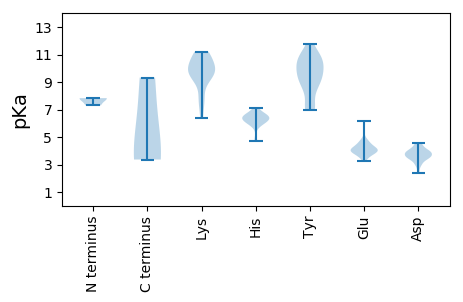
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7CL82|A0A0A7CL82_9VIRU Replication-associated protein OS=Sewage-associated gemycircularvirus-10b OX=1519410 PE=3 SV=1
MM1 pKa = 7.83SSFRR5 pKa = 11.84FQARR9 pKa = 11.84YY10 pKa = 9.86ALLTYY15 pKa = 7.21PQCGDD20 pKa = 3.86LDD22 pKa = 3.85PFAVVNHH29 pKa = 6.38LAGLGAEE36 pKa = 4.46CIIGRR41 pKa = 11.84EE42 pKa = 3.88NHH44 pKa = 6.16TDD46 pKa = 3.34GGLHH50 pKa = 6.08LHH52 pKa = 6.6AFVDD56 pKa = 4.51FGVKK60 pKa = 9.88YY61 pKa = 8.61RR62 pKa = 11.84TRR64 pKa = 11.84NARR67 pKa = 11.84AFDD70 pKa = 3.89VEE72 pKa = 4.6GCHH75 pKa = 6.51PNVSPSRR82 pKa = 11.84GTPEE86 pKa = 3.58EE87 pKa = 4.81GYY89 pKa = 10.84DD90 pKa = 3.63YY91 pKa = 10.79AIKK94 pKa = 10.96DD95 pKa = 3.59GDD97 pKa = 4.01VVAGGLEE104 pKa = 4.09RR105 pKa = 11.84PSGSRR110 pKa = 11.84VDD112 pKa = 3.91ANGGVWAEE120 pKa = 4.11IINAKK125 pKa = 9.97SEE127 pKa = 4.02QEE129 pKa = 3.88FWDD132 pKa = 3.5LCEE135 pKa = 4.08RR136 pKa = 11.84LAPRR140 pKa = 11.84SLVTSFTQLRR150 pKa = 11.84AYY152 pKa = 9.55AAWKK156 pKa = 9.13FPPIRR161 pKa = 11.84VPYY164 pKa = 6.94EE165 pKa = 3.74TPEE168 pKa = 4.05GVNIDD173 pKa = 3.83TSWVVEE179 pKa = 4.03LDD181 pKa = 2.98QWVQQNLGRR190 pKa = 11.84SEE192 pKa = 4.28TGGKK196 pKa = 9.33
MM1 pKa = 7.83SSFRR5 pKa = 11.84FQARR9 pKa = 11.84YY10 pKa = 9.86ALLTYY15 pKa = 7.21PQCGDD20 pKa = 3.86LDD22 pKa = 3.85PFAVVNHH29 pKa = 6.38LAGLGAEE36 pKa = 4.46CIIGRR41 pKa = 11.84EE42 pKa = 3.88NHH44 pKa = 6.16TDD46 pKa = 3.34GGLHH50 pKa = 6.08LHH52 pKa = 6.6AFVDD56 pKa = 4.51FGVKK60 pKa = 9.88YY61 pKa = 8.61RR62 pKa = 11.84TRR64 pKa = 11.84NARR67 pKa = 11.84AFDD70 pKa = 3.89VEE72 pKa = 4.6GCHH75 pKa = 6.51PNVSPSRR82 pKa = 11.84GTPEE86 pKa = 3.58EE87 pKa = 4.81GYY89 pKa = 10.84DD90 pKa = 3.63YY91 pKa = 10.79AIKK94 pKa = 10.96DD95 pKa = 3.59GDD97 pKa = 4.01VVAGGLEE104 pKa = 4.09RR105 pKa = 11.84PSGSRR110 pKa = 11.84VDD112 pKa = 3.91ANGGVWAEE120 pKa = 4.11IINAKK125 pKa = 9.97SEE127 pKa = 4.02QEE129 pKa = 3.88FWDD132 pKa = 3.5LCEE135 pKa = 4.08RR136 pKa = 11.84LAPRR140 pKa = 11.84SLVTSFTQLRR150 pKa = 11.84AYY152 pKa = 9.55AAWKK156 pKa = 9.13FPPIRR161 pKa = 11.84VPYY164 pKa = 6.94EE165 pKa = 3.74TPEE168 pKa = 4.05GVNIDD173 pKa = 3.83TSWVVEE179 pKa = 4.03LDD181 pKa = 2.98QWVQQNLGRR190 pKa = 11.84SEE192 pKa = 4.28TGGKK196 pKa = 9.33
Molecular weight: 21.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7CL99|A0A0A7CL99_9VIRU Capsid protein OS=Sewage-associated gemycircularvirus-10b OX=1519410 PE=4 SV=1
MM1 pKa = 7.3SLARR5 pKa = 11.84RR6 pKa = 11.84PVRR9 pKa = 11.84RR10 pKa = 11.84SRR12 pKa = 11.84KK13 pKa = 6.36TYY15 pKa = 9.4RR16 pKa = 11.84RR17 pKa = 11.84GGRR20 pKa = 11.84VTRR23 pKa = 11.84SRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84STRR30 pKa = 11.84TSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84VGRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84MTTRR45 pKa = 11.84RR46 pKa = 11.84VLNIASRR53 pKa = 11.84KK54 pKa = 9.62KK55 pKa = 9.56VDD57 pKa = 3.09NMLPVVVAEE66 pKa = 4.69DD67 pKa = 3.42NVTTVGPYY75 pKa = 9.07TSPSPLLCLFVPNARR90 pKa = 11.84TTRR93 pKa = 11.84TPVTNPAVRR102 pKa = 11.84NSSDD106 pKa = 2.87IFAVGYY112 pKa = 9.14KK113 pKa = 9.64EE114 pKa = 4.03RR115 pKa = 11.84VRR117 pKa = 11.84IDD119 pKa = 3.09ILGGGTFMWRR129 pKa = 11.84RR130 pKa = 11.84IVFMLKK136 pKa = 10.54GSDD139 pKa = 3.5LRR141 pKa = 11.84VAMDD145 pKa = 3.85SSNAGNIPNQLFDD158 pKa = 3.24QTTEE162 pKa = 4.13GGCRR166 pKa = 11.84RR167 pKa = 11.84VIGPLLGVTNAQDD180 pKa = 3.51EE181 pKa = 4.51LQSYY185 pKa = 7.05VFRR188 pKa = 11.84GQEE191 pKa = 3.75DD192 pKa = 4.44TDD194 pKa = 3.49WADD197 pKa = 3.34QFTAPLDD204 pKa = 3.58TRR206 pKa = 11.84RR207 pKa = 11.84ITVHH211 pKa = 5.82SDD213 pKa = 2.41KK214 pKa = 10.97LRR216 pKa = 11.84TIRR219 pKa = 11.84PGNDD223 pKa = 2.46TGASRR228 pKa = 11.84LYY230 pKa = 10.16KK231 pKa = 9.37CWYY234 pKa = 8.3PIRR237 pKa = 11.84RR238 pKa = 11.84TIAYY242 pKa = 9.2EE243 pKa = 4.32DD244 pKa = 4.14DD245 pKa = 4.25LEE247 pKa = 6.16SDD249 pKa = 4.19VVGDD253 pKa = 4.43RR254 pKa = 11.84PFSTAGLRR262 pKa = 11.84GVGDD266 pKa = 4.13MYY268 pKa = 11.73VMDD271 pKa = 4.6IMGITNLTPDD281 pKa = 4.03APEE284 pKa = 3.46TSYY287 pKa = 11.17RR288 pKa = 11.84FSPEE292 pKa = 3.23GSFYY296 pKa = 9.4WHH298 pKa = 6.64EE299 pKa = 4.04RR300 pKa = 3.36
MM1 pKa = 7.3SLARR5 pKa = 11.84RR6 pKa = 11.84PVRR9 pKa = 11.84RR10 pKa = 11.84SRR12 pKa = 11.84KK13 pKa = 6.36TYY15 pKa = 9.4RR16 pKa = 11.84RR17 pKa = 11.84GGRR20 pKa = 11.84VTRR23 pKa = 11.84SRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84STRR30 pKa = 11.84TSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84VGRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84MTTRR45 pKa = 11.84RR46 pKa = 11.84VLNIASRR53 pKa = 11.84KK54 pKa = 9.62KK55 pKa = 9.56VDD57 pKa = 3.09NMLPVVVAEE66 pKa = 4.69DD67 pKa = 3.42NVTTVGPYY75 pKa = 9.07TSPSPLLCLFVPNARR90 pKa = 11.84TTRR93 pKa = 11.84TPVTNPAVRR102 pKa = 11.84NSSDD106 pKa = 2.87IFAVGYY112 pKa = 9.14KK113 pKa = 9.64EE114 pKa = 4.03RR115 pKa = 11.84VRR117 pKa = 11.84IDD119 pKa = 3.09ILGGGTFMWRR129 pKa = 11.84RR130 pKa = 11.84IVFMLKK136 pKa = 10.54GSDD139 pKa = 3.5LRR141 pKa = 11.84VAMDD145 pKa = 3.85SSNAGNIPNQLFDD158 pKa = 3.24QTTEE162 pKa = 4.13GGCRR166 pKa = 11.84RR167 pKa = 11.84VIGPLLGVTNAQDD180 pKa = 3.51EE181 pKa = 4.51LQSYY185 pKa = 7.05VFRR188 pKa = 11.84GQEE191 pKa = 3.75DD192 pKa = 4.44TDD194 pKa = 3.49WADD197 pKa = 3.34QFTAPLDD204 pKa = 3.58TRR206 pKa = 11.84RR207 pKa = 11.84ITVHH211 pKa = 5.82SDD213 pKa = 2.41KK214 pKa = 10.97LRR216 pKa = 11.84TIRR219 pKa = 11.84PGNDD223 pKa = 2.46TGASRR228 pKa = 11.84LYY230 pKa = 10.16KK231 pKa = 9.37CWYY234 pKa = 8.3PIRR237 pKa = 11.84RR238 pKa = 11.84TIAYY242 pKa = 9.2EE243 pKa = 4.32DD244 pKa = 4.14DD245 pKa = 4.25LEE247 pKa = 6.16SDD249 pKa = 4.19VVGDD253 pKa = 4.43RR254 pKa = 11.84PFSTAGLRR262 pKa = 11.84GVGDD266 pKa = 4.13MYY268 pKa = 11.73VMDD271 pKa = 4.6IMGITNLTPDD281 pKa = 4.03APEE284 pKa = 3.46TSYY287 pKa = 11.17RR288 pKa = 11.84FSPEE292 pKa = 3.23GSFYY296 pKa = 9.4WHH298 pKa = 6.64EE299 pKa = 4.04RR300 pKa = 3.36
Molecular weight: 34.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
807 |
196 |
311 |
269.0 |
30.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.691 ± 0.833 | 1.487 ± 0.237 |
6.815 ± 0.24 | 4.957 ± 0.853 |
4.461 ± 0.551 | 8.922 ± 0.59 |
1.859 ± 0.531 | 3.965 ± 0.175 |
3.222 ± 0.624 | 7.311 ± 0.533 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.487 ± 0.673 | 4.213 ± 0.141 |
5.452 ± 0.15 | 2.726 ± 0.357 |
9.789 ± 2.333 | 6.444 ± 0.311 |
5.948 ± 1.537 | 8.055 ± 0.167 |
2.354 ± 0.487 | 3.841 ± 0.324 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
