
Leptonychotes weddellii papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
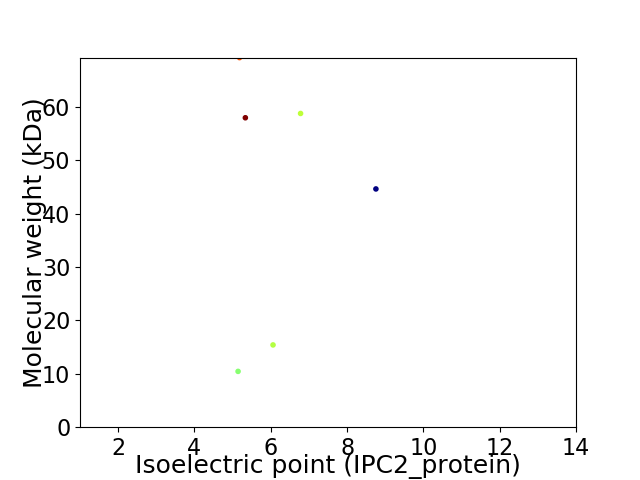
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
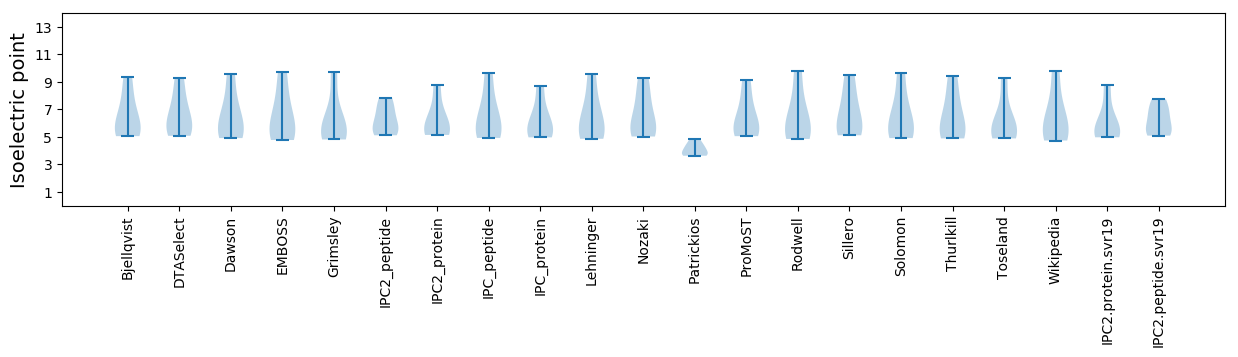
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I8B2N0|A0A2I8B2N0_9PAPI Major capsid protein L1 OS=Leptonychotes weddellii papillomavirus 2 OX=2077303 GN=L1 PE=3 SV=1
MM1 pKa = 7.96RR2 pKa = 11.84GQEE5 pKa = 4.13PAPLDD10 pKa = 3.55LAINLHH16 pKa = 5.85EE17 pKa = 4.45LVIPEE22 pKa = 4.09NLLSGEE28 pKa = 4.08EE29 pKa = 4.45LSPNWEE35 pKa = 4.06EE36 pKa = 4.0EE37 pKa = 4.2EE38 pKa = 4.33EE39 pKa = 4.03PRR41 pKa = 11.84NPYY44 pKa = 10.13KK45 pKa = 10.03VTCSCVSCGASLRR58 pKa = 11.84LAVVATNPAIRR69 pKa = 11.84GLQTLLLGDD78 pKa = 4.47LSILCAQCAKK88 pKa = 9.67STLRR92 pKa = 11.84HH93 pKa = 5.25GRR95 pKa = 11.84QQ96 pKa = 2.98
MM1 pKa = 7.96RR2 pKa = 11.84GQEE5 pKa = 4.13PAPLDD10 pKa = 3.55LAINLHH16 pKa = 5.85EE17 pKa = 4.45LVIPEE22 pKa = 4.09NLLSGEE28 pKa = 4.08EE29 pKa = 4.45LSPNWEE35 pKa = 4.06EE36 pKa = 4.0EE37 pKa = 4.2EE38 pKa = 4.33EE39 pKa = 4.03PRR41 pKa = 11.84NPYY44 pKa = 10.13KK45 pKa = 10.03VTCSCVSCGASLRR58 pKa = 11.84LAVVATNPAIRR69 pKa = 11.84GLQTLLLGDD78 pKa = 4.47LSILCAQCAKK88 pKa = 9.67STLRR92 pKa = 11.84HH93 pKa = 5.25GRR95 pKa = 11.84QQ96 pKa = 2.98
Molecular weight: 10.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I8B2M5|A0A2I8B2M5_9PAPI Minor capsid protein L2 OS=Leptonychotes weddellii papillomavirus 2 OX=2077303 GN=L2 PE=3 SV=1
MM1 pKa = 6.92ATLRR5 pKa = 11.84EE6 pKa = 4.13RR7 pKa = 11.84LASLQEE13 pKa = 4.29TLSEE17 pKa = 4.36LYY19 pKa = 10.56EE20 pKa = 4.76KK21 pKa = 10.77DD22 pKa = 3.28STEE25 pKa = 4.17LQDD28 pKa = 6.06QITFWDD34 pKa = 3.71LTRR37 pKa = 11.84QEE39 pKa = 4.66NLLMHH44 pKa = 6.3YY45 pKa = 9.74GKK47 pKa = 10.42KK48 pKa = 9.97RR49 pKa = 11.84GLHH52 pKa = 5.87SIGLQTLPASQVSEE66 pKa = 4.31INTKK70 pKa = 9.41HH71 pKa = 6.64AIMMKK76 pKa = 10.42LVLTSLSKK84 pKa = 10.79SPYY87 pKa = 10.73AKK89 pKa = 10.52DD90 pKa = 3.0PWTMRR95 pKa = 11.84EE96 pKa = 3.87SSYY99 pKa = 10.89EE100 pKa = 4.0LYY102 pKa = 10.17MADD105 pKa = 4.28PSYY108 pKa = 10.1TLKK111 pKa = 10.65KK112 pKa = 10.08KK113 pKa = 9.73PVSVEE118 pKa = 3.82VFFDD122 pKa = 3.72NDD124 pKa = 3.51PEE126 pKa = 4.18NYY128 pKa = 9.75YY129 pKa = 10.05PYY131 pKa = 10.32TLWQEE136 pKa = 4.07VYY138 pKa = 10.82YY139 pKa = 10.55EE140 pKa = 4.88DD141 pKa = 6.08EE142 pKa = 4.54DD143 pKa = 4.03NQWYY147 pKa = 9.85KK148 pKa = 11.25GRR150 pKa = 11.84GGSDD154 pKa = 3.23DD155 pKa = 3.42TGLFYY160 pKa = 10.98VDD162 pKa = 2.83HH163 pKa = 7.65RR164 pKa = 11.84GRR166 pKa = 11.84KK167 pKa = 7.77VYY169 pKa = 9.82YY170 pKa = 10.5VKK172 pKa = 10.81FEE174 pKa = 4.1EE175 pKa = 4.5DD176 pKa = 2.83AARR179 pKa = 11.84YY180 pKa = 7.02GTTGQWEE187 pKa = 4.39VRR189 pKa = 11.84NKK191 pKa = 8.17QTVSALSSKK200 pKa = 10.13PPWGDD205 pKa = 2.72ISSSSPEE212 pKa = 3.97HH213 pKa = 6.53GEE215 pKa = 4.32SPGLGDD221 pKa = 4.27ASTSEE226 pKa = 4.53TPRR229 pKa = 11.84KK230 pKa = 8.99RR231 pKa = 11.84QRR233 pKa = 11.84EE234 pKa = 3.97RR235 pKa = 11.84QVRR238 pKa = 11.84FGGDD242 pKa = 2.69SAANNRR248 pKa = 11.84GKK250 pKa = 10.37RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84QGEE256 pKa = 4.03RR257 pKa = 11.84SPQTRR262 pKa = 11.84SQRR265 pKa = 11.84TNGDD269 pKa = 3.2IASGGPAFISPDD281 pKa = 3.32EE282 pKa = 4.27VGSRR286 pKa = 11.84DD287 pKa = 3.36RR288 pKa = 11.84TVGRR292 pKa = 11.84GLQTRR297 pKa = 11.84LSRR300 pKa = 11.84LQEE303 pKa = 3.95EE304 pKa = 4.55ALDD307 pKa = 3.98PPVILLKK314 pKa = 10.93GCARR318 pKa = 11.84NLKK321 pKa = 9.63SFRR324 pKa = 11.84NRR326 pKa = 11.84HH327 pKa = 5.51RR328 pKa = 11.84DD329 pKa = 3.27VALKK333 pKa = 9.29FHH335 pKa = 6.78AQTSSVFKK343 pKa = 10.51WLGDD347 pKa = 3.51TSGKK351 pKa = 10.05RR352 pKa = 11.84NQSRR356 pKa = 11.84MLLAFDD362 pKa = 4.13NEE364 pKa = 4.38EE365 pKa = 3.88NRR367 pKa = 11.84TKK369 pKa = 11.16YY370 pKa = 10.48VMSVSFPKK378 pKa = 9.84GTRR381 pKa = 11.84FAYY384 pKa = 10.45GNLNSLL390 pKa = 4.17
MM1 pKa = 6.92ATLRR5 pKa = 11.84EE6 pKa = 4.13RR7 pKa = 11.84LASLQEE13 pKa = 4.29TLSEE17 pKa = 4.36LYY19 pKa = 10.56EE20 pKa = 4.76KK21 pKa = 10.77DD22 pKa = 3.28STEE25 pKa = 4.17LQDD28 pKa = 6.06QITFWDD34 pKa = 3.71LTRR37 pKa = 11.84QEE39 pKa = 4.66NLLMHH44 pKa = 6.3YY45 pKa = 9.74GKK47 pKa = 10.42KK48 pKa = 9.97RR49 pKa = 11.84GLHH52 pKa = 5.87SIGLQTLPASQVSEE66 pKa = 4.31INTKK70 pKa = 9.41HH71 pKa = 6.64AIMMKK76 pKa = 10.42LVLTSLSKK84 pKa = 10.79SPYY87 pKa = 10.73AKK89 pKa = 10.52DD90 pKa = 3.0PWTMRR95 pKa = 11.84EE96 pKa = 3.87SSYY99 pKa = 10.89EE100 pKa = 4.0LYY102 pKa = 10.17MADD105 pKa = 4.28PSYY108 pKa = 10.1TLKK111 pKa = 10.65KK112 pKa = 10.08KK113 pKa = 9.73PVSVEE118 pKa = 3.82VFFDD122 pKa = 3.72NDD124 pKa = 3.51PEE126 pKa = 4.18NYY128 pKa = 9.75YY129 pKa = 10.05PYY131 pKa = 10.32TLWQEE136 pKa = 4.07VYY138 pKa = 10.82YY139 pKa = 10.55EE140 pKa = 4.88DD141 pKa = 6.08EE142 pKa = 4.54DD143 pKa = 4.03NQWYY147 pKa = 9.85KK148 pKa = 11.25GRR150 pKa = 11.84GGSDD154 pKa = 3.23DD155 pKa = 3.42TGLFYY160 pKa = 10.98VDD162 pKa = 2.83HH163 pKa = 7.65RR164 pKa = 11.84GRR166 pKa = 11.84KK167 pKa = 7.77VYY169 pKa = 9.82YY170 pKa = 10.5VKK172 pKa = 10.81FEE174 pKa = 4.1EE175 pKa = 4.5DD176 pKa = 2.83AARR179 pKa = 11.84YY180 pKa = 7.02GTTGQWEE187 pKa = 4.39VRR189 pKa = 11.84NKK191 pKa = 8.17QTVSALSSKK200 pKa = 10.13PPWGDD205 pKa = 2.72ISSSSPEE212 pKa = 3.97HH213 pKa = 6.53GEE215 pKa = 4.32SPGLGDD221 pKa = 4.27ASTSEE226 pKa = 4.53TPRR229 pKa = 11.84KK230 pKa = 8.99RR231 pKa = 11.84QRR233 pKa = 11.84EE234 pKa = 3.97RR235 pKa = 11.84QVRR238 pKa = 11.84FGGDD242 pKa = 2.69SAANNRR248 pKa = 11.84GKK250 pKa = 10.37RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84QGEE256 pKa = 4.03RR257 pKa = 11.84SPQTRR262 pKa = 11.84SQRR265 pKa = 11.84TNGDD269 pKa = 3.2IASGGPAFISPDD281 pKa = 3.32EE282 pKa = 4.27VGSRR286 pKa = 11.84DD287 pKa = 3.36RR288 pKa = 11.84TVGRR292 pKa = 11.84GLQTRR297 pKa = 11.84LSRR300 pKa = 11.84LQEE303 pKa = 3.95EE304 pKa = 4.55ALDD307 pKa = 3.98PPVILLKK314 pKa = 10.93GCARR318 pKa = 11.84NLKK321 pKa = 9.63SFRR324 pKa = 11.84NRR326 pKa = 11.84HH327 pKa = 5.51RR328 pKa = 11.84DD329 pKa = 3.27VALKK333 pKa = 9.29FHH335 pKa = 6.78AQTSSVFKK343 pKa = 10.51WLGDD347 pKa = 3.51TSGKK351 pKa = 10.05RR352 pKa = 11.84NQSRR356 pKa = 11.84MLLAFDD362 pKa = 4.13NEE364 pKa = 4.38EE365 pKa = 3.88NRR367 pKa = 11.84TKK369 pKa = 11.16YY370 pKa = 10.48VMSVSFPKK378 pKa = 9.84GTRR381 pKa = 11.84FAYY384 pKa = 10.45GNLNSLL390 pKa = 4.17
Molecular weight: 44.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2297 |
96 |
618 |
382.8 |
42.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.485 ± 0.469 | 2.264 ± 0.791 |
5.834 ± 0.282 | 6.487 ± 0.433 |
3.875 ± 0.508 | 7.357 ± 0.847 |
1.741 ± 0.202 | 4.092 ± 0.79 |
5.355 ± 0.946 | 9.099 ± 0.737 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.611 ± 0.439 | 4.484 ± 0.659 |
5.921 ± 0.761 | 4.005 ± 0.316 |
6.051 ± 0.798 | 8.663 ± 0.82 |
6.661 ± 0.57 | 6.226 ± 0.459 |
1.35 ± 0.363 | 3.439 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
