
Diaminobutyricibacter tongyongensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Diaminobutyricibacter
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
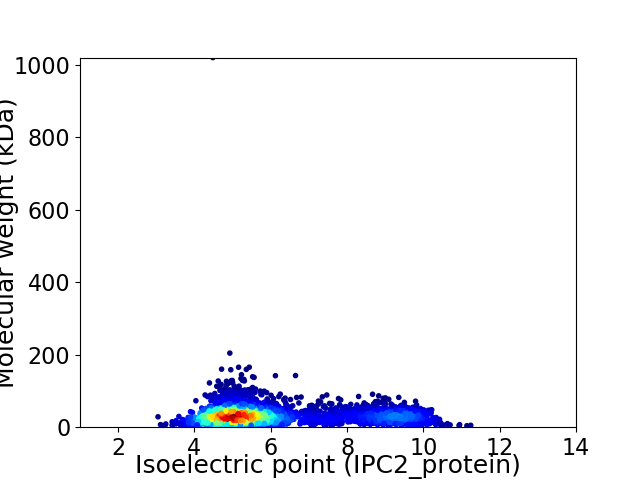
Virtual 2D-PAGE plot for 3799 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
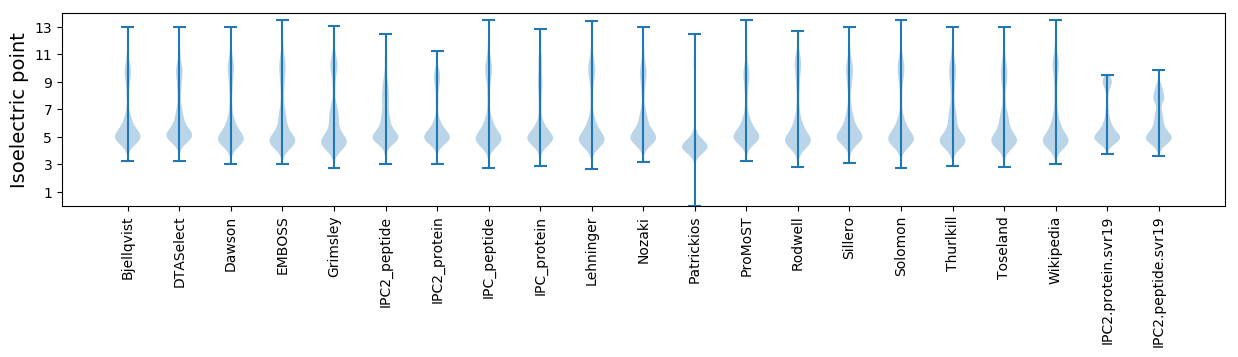
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L9XSM7|A0A6L9XSM7_9MICO Response regulator transcription factor OS=Diaminobutyricibacter tongyongensis OX=1268043 GN=G3T36_00865 PE=4 SV=1
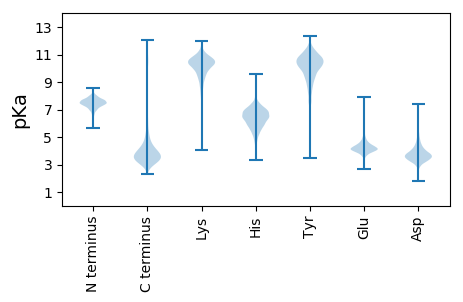
MM1 pKa = 7.84DD2 pKa = 3.86TTDD5 pKa = 3.54GATGGTGIDD14 pKa = 3.6QGNVQQPSAAHH25 pKa = 5.86SGVSRR30 pKa = 11.84GRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84VVVLSAALAATVVAGATAGVAANGFASRR63 pKa = 11.84SAAAASTSSTSANIHH78 pKa = 5.86NALGPGSYY86 pKa = 10.58GYY88 pKa = 10.64GGYY91 pKa = 10.92GSGGYY96 pKa = 9.59GGYY99 pKa = 10.35GGYY102 pKa = 10.94ASGGYY107 pKa = 10.24GSGGYY112 pKa = 9.86GSSSGSTGSAALASATPAEE131 pKa = 4.41EE132 pKa = 4.11VGVVDD137 pKa = 3.51ITSEE141 pKa = 3.9LTYY144 pKa = 10.61QQAEE148 pKa = 4.28SAGTGLVLTSDD159 pKa = 4.83GEE161 pKa = 4.28ILTNNHH167 pKa = 4.8VVAGATSISVTVVSTGATFTATVVGTDD194 pKa = 3.25ATDD197 pKa = 4.88DD198 pKa = 3.57IAVLHH203 pKa = 6.53LSNASGLSTAHH214 pKa = 7.36LDD216 pKa = 3.85TSNSPAVGDD225 pKa = 3.7AVTGVGNAGGTGGTPSASPGTVTALDD251 pKa = 3.59QTITTQAEE259 pKa = 4.24GSAASEE265 pKa = 4.16TLNGLIEE272 pKa = 4.27TDD274 pKa = 3.59ADD276 pKa = 3.55IQAGDD281 pKa = 3.23SGGPLFNSADD291 pKa = 3.58QVIGIDD297 pKa = 3.32TAAASGGATPAGYY310 pKa = 10.33AIPIRR315 pKa = 11.84SALAVAGEE323 pKa = 4.32IVAGHH328 pKa = 7.03ASSTVTIGYY337 pKa = 7.9PAFLGVEE344 pKa = 4.42VGSSADD350 pKa = 3.31GTAAATPGAAIAGVIDD366 pKa = 4.3GSPAATAGLAAGDD379 pKa = 4.44TITALNGTTIDD390 pKa = 3.63SQQALTATLQTLAPGQRR407 pKa = 11.84VSIAWTDD414 pKa = 3.29QSGASQSAPITLASGPVAA432 pKa = 4.2
MM1 pKa = 7.84DD2 pKa = 3.86TTDD5 pKa = 3.54GATGGTGIDD14 pKa = 3.6QGNVQQPSAAHH25 pKa = 5.86SGVSRR30 pKa = 11.84GRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84VVVLSAALAATVVAGATAGVAANGFASRR63 pKa = 11.84SAAAASTSSTSANIHH78 pKa = 5.86NALGPGSYY86 pKa = 10.58GYY88 pKa = 10.64GGYY91 pKa = 10.92GSGGYY96 pKa = 9.59GGYY99 pKa = 10.35GGYY102 pKa = 10.94ASGGYY107 pKa = 10.24GSGGYY112 pKa = 9.86GSSSGSTGSAALASATPAEE131 pKa = 4.41EE132 pKa = 4.11VGVVDD137 pKa = 3.51ITSEE141 pKa = 3.9LTYY144 pKa = 10.61QQAEE148 pKa = 4.28SAGTGLVLTSDD159 pKa = 4.83GEE161 pKa = 4.28ILTNNHH167 pKa = 4.8VVAGATSISVTVVSTGATFTATVVGTDD194 pKa = 3.25ATDD197 pKa = 4.88DD198 pKa = 3.57IAVLHH203 pKa = 6.53LSNASGLSTAHH214 pKa = 7.36LDD216 pKa = 3.85TSNSPAVGDD225 pKa = 3.7AVTGVGNAGGTGGTPSASPGTVTALDD251 pKa = 3.59QTITTQAEE259 pKa = 4.24GSAASEE265 pKa = 4.16TLNGLIEE272 pKa = 4.27TDD274 pKa = 3.59ADD276 pKa = 3.55IQAGDD281 pKa = 3.23SGGPLFNSADD291 pKa = 3.58QVIGIDD297 pKa = 3.32TAAASGGATPAGYY310 pKa = 10.33AIPIRR315 pKa = 11.84SALAVAGEE323 pKa = 4.32IVAGHH328 pKa = 7.03ASSTVTIGYY337 pKa = 7.9PAFLGVEE344 pKa = 4.42VGSSADD350 pKa = 3.31GTAAATPGAAIAGVIDD366 pKa = 4.3GSPAATAGLAAGDD379 pKa = 4.44TITALNGTTIDD390 pKa = 3.63SQQALTATLQTLAPGQRR407 pKa = 11.84VSIAWTDD414 pKa = 3.29QSGASQSAPITLASGPVAA432 pKa = 4.2
Molecular weight: 40.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L9XVF8|A0A6L9XVF8_9MICO Alpha/beta hydrolase OS=Diaminobutyricibacter tongyongensis OX=1268043 GN=G3T36_05890 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.46THH17 pKa = 5.11GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.79GRR40 pKa = 11.84TKK42 pKa = 10.84LSAA45 pKa = 3.61
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.46THH17 pKa = 5.11GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.79GRR40 pKa = 11.84TKK42 pKa = 10.84LSAA45 pKa = 3.61
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1213229 |
27 |
10786 |
319.4 |
34.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.001 ± 0.075 | 0.493 ± 0.009 |
6.007 ± 0.038 | 5.36 ± 0.053 |
3.321 ± 0.027 | 8.801 ± 0.056 |
1.976 ± 0.018 | 4.87 ± 0.031 |
2.199 ± 0.029 | 10.032 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.736 ± 0.02 | 2.328 ± 0.031 |
5.337 ± 0.033 | 2.846 ± 0.025 |
6.919 ± 0.058 | 6.06 ± 0.046 |
6.268 ± 0.074 | 8.813 ± 0.039 |
1.511 ± 0.023 | 2.122 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
