
Tetradesmus obliquus (Green alga) (Acutodesmus obliquus)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Chlorophyceae; CS clade; Sphaeropleales; Scenedesmaceae; Tetradesmus
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
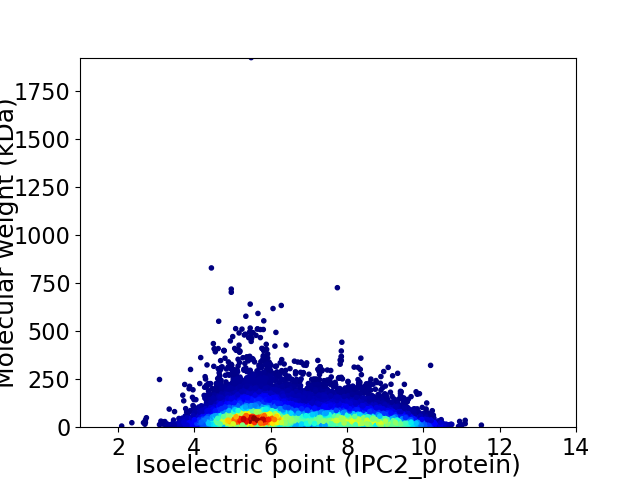
Virtual 2D-PAGE plot for 18538 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A383VB91|A0A383VB91_TETOB MIT domain-containing protein OS=Tetradesmus obliquus OX=3088 GN=BQ4739_LOCUS3053 PE=4 SV=1
MM1 pKa = 7.54ALSDD5 pKa = 4.82DD6 pKa = 3.94PHH8 pKa = 9.03ASDD11 pKa = 4.86QEE13 pKa = 4.39SDD15 pKa = 3.9GEE17 pKa = 4.35GLDD20 pKa = 3.55EE21 pKa = 5.66EE22 pKa = 4.89QYY24 pKa = 11.03QWLQEE29 pKa = 4.05QLAHH33 pKa = 6.78IGNLHH38 pKa = 6.26IEE40 pKa = 4.29GDD42 pKa = 3.8EE43 pKa = 5.1EE44 pKa = 4.0EE45 pKa = 5.79DD46 pKa = 3.25EE47 pKa = 4.37WADD50 pKa = 3.45PSGGLLVNIFAACEE64 pKa = 3.9EE65 pKa = 4.38GSMDD69 pKa = 4.65QLTTNIQEE77 pKa = 4.68LKK79 pKa = 9.67DD80 pKa = 3.46TQYY83 pKa = 11.81DD84 pKa = 4.01VNTPGPDD91 pKa = 3.3GDD93 pKa = 4.22TALHH97 pKa = 6.39LACLYY102 pKa = 10.12GHH104 pKa = 5.61QQCAEE109 pKa = 4.25LLLQEE114 pKa = 4.44GAKK117 pKa = 10.16ADD119 pKa = 3.67AVNPEE124 pKa = 4.72DD125 pKa = 4.04GTTPLHH131 pKa = 6.66DD132 pKa = 4.02AAAGGYY138 pKa = 10.05AGIVQMLLEE147 pKa = 4.19KK148 pKa = 10.46SGPGSVSLQDD158 pKa = 3.58SDD160 pKa = 5.31GDD162 pKa = 4.17TALHH166 pKa = 5.44NAARR170 pKa = 11.84GGHH173 pKa = 6.15LAVVQQLLAAGAAPSVLNSSGKK195 pKa = 8.01TPAGEE200 pKa = 4.07SDD202 pKa = 3.34NQEE205 pKa = 3.78VVRR208 pKa = 11.84ALVAAAAAALGVGGTAPAAAAAAPAAAPAADD239 pKa = 3.74TAACGASMDD248 pKa = 3.87AQQ250 pKa = 3.49
MM1 pKa = 7.54ALSDD5 pKa = 4.82DD6 pKa = 3.94PHH8 pKa = 9.03ASDD11 pKa = 4.86QEE13 pKa = 4.39SDD15 pKa = 3.9GEE17 pKa = 4.35GLDD20 pKa = 3.55EE21 pKa = 5.66EE22 pKa = 4.89QYY24 pKa = 11.03QWLQEE29 pKa = 4.05QLAHH33 pKa = 6.78IGNLHH38 pKa = 6.26IEE40 pKa = 4.29GDD42 pKa = 3.8EE43 pKa = 5.1EE44 pKa = 4.0EE45 pKa = 5.79DD46 pKa = 3.25EE47 pKa = 4.37WADD50 pKa = 3.45PSGGLLVNIFAACEE64 pKa = 3.9EE65 pKa = 4.38GSMDD69 pKa = 4.65QLTTNIQEE77 pKa = 4.68LKK79 pKa = 9.67DD80 pKa = 3.46TQYY83 pKa = 11.81DD84 pKa = 4.01VNTPGPDD91 pKa = 3.3GDD93 pKa = 4.22TALHH97 pKa = 6.39LACLYY102 pKa = 10.12GHH104 pKa = 5.61QQCAEE109 pKa = 4.25LLLQEE114 pKa = 4.44GAKK117 pKa = 10.16ADD119 pKa = 3.67AVNPEE124 pKa = 4.72DD125 pKa = 4.04GTTPLHH131 pKa = 6.66DD132 pKa = 4.02AAAGGYY138 pKa = 10.05AGIVQMLLEE147 pKa = 4.19KK148 pKa = 10.46SGPGSVSLQDD158 pKa = 3.58SDD160 pKa = 5.31GDD162 pKa = 4.17TALHH166 pKa = 5.44NAARR170 pKa = 11.84GGHH173 pKa = 6.15LAVVQQLLAAGAAPSVLNSSGKK195 pKa = 8.01TPAGEE200 pKa = 4.07SDD202 pKa = 3.34NQEE205 pKa = 3.78VVRR208 pKa = 11.84ALVAAAAAALGVGGTAPAAAAAAPAAAPAADD239 pKa = 3.74TAACGASMDD248 pKa = 3.87AQQ250 pKa = 3.49
Molecular weight: 25.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A383WP31|A0A383WP31_TETOB Uncharacterized protein OS=Tetradesmus obliquus OX=3088 GN=BQ4739_LOCUS19502 PE=4 SV=1
MM1 pKa = 7.36ARR3 pKa = 11.84SHH5 pKa = 7.12VITTLVLIAFAAVVTANVEE24 pKa = 4.0DD25 pKa = 3.86DD26 pKa = 3.59HH27 pKa = 6.44TRR29 pKa = 11.84KK30 pKa = 9.9LLQGNARR37 pKa = 11.84STGAMMSQHH46 pKa = 5.68STSAAIKK53 pKa = 7.41TTVQNAQAGRR63 pKa = 11.84GSTGNTKK70 pKa = 10.3AATRR74 pKa = 11.84TGSVATTVLARR85 pKa = 11.84PSGPNTATAGAVTRR99 pKa = 11.84AATPANRR106 pKa = 11.84PTGRR110 pKa = 11.84HH111 pKa = 5.43LLQARR116 pKa = 11.84NTGAAGSRR124 pKa = 11.84ATASQATTSAAIKK137 pKa = 8.85QTVQAANRR145 pKa = 11.84GAPVAAKK152 pKa = 9.19PATRR156 pKa = 11.84TGQTAAQVIARR167 pKa = 11.84PAGLPATVTSQAVTTRR183 pKa = 11.84PAQQGARR190 pKa = 11.84TGRR193 pKa = 11.84HH194 pKa = 5.36LLQARR199 pKa = 11.84NTGAAGSRR207 pKa = 11.84ATASQATTSAAIKK220 pKa = 8.85QTVQAANRR228 pKa = 11.84GTPVAAKK235 pKa = 9.01PATRR239 pKa = 11.84TGQTAAQVIARR250 pKa = 11.84PAGLPATVTSQAVTTRR266 pKa = 11.84PAQQRR271 pKa = 11.84TGRR274 pKa = 11.84HH275 pKa = 5.18LLQNSRR281 pKa = 11.84TGANQGRR288 pKa = 11.84ATAAQGTTSAAIRR301 pKa = 11.84QTVQAANRR309 pKa = 11.84GAPVAAKK316 pKa = 9.19PATRR320 pKa = 11.84TGQAAAQTIARR331 pKa = 11.84PAGLPATVTSQAVTTRR347 pKa = 11.84PAQQNPRR354 pKa = 11.84GLL356 pKa = 3.62
MM1 pKa = 7.36ARR3 pKa = 11.84SHH5 pKa = 7.12VITTLVLIAFAAVVTANVEE24 pKa = 4.0DD25 pKa = 3.86DD26 pKa = 3.59HH27 pKa = 6.44TRR29 pKa = 11.84KK30 pKa = 9.9LLQGNARR37 pKa = 11.84STGAMMSQHH46 pKa = 5.68STSAAIKK53 pKa = 7.41TTVQNAQAGRR63 pKa = 11.84GSTGNTKK70 pKa = 10.3AATRR74 pKa = 11.84TGSVATTVLARR85 pKa = 11.84PSGPNTATAGAVTRR99 pKa = 11.84AATPANRR106 pKa = 11.84PTGRR110 pKa = 11.84HH111 pKa = 5.43LLQARR116 pKa = 11.84NTGAAGSRR124 pKa = 11.84ATASQATTSAAIKK137 pKa = 8.85QTVQAANRR145 pKa = 11.84GAPVAAKK152 pKa = 9.19PATRR156 pKa = 11.84TGQTAAQVIARR167 pKa = 11.84PAGLPATVTSQAVTTRR183 pKa = 11.84PAQQGARR190 pKa = 11.84TGRR193 pKa = 11.84HH194 pKa = 5.36LLQARR199 pKa = 11.84NTGAAGSRR207 pKa = 11.84ATASQATTSAAIKK220 pKa = 8.85QTVQAANRR228 pKa = 11.84GTPVAAKK235 pKa = 9.01PATRR239 pKa = 11.84TGQTAAQVIARR250 pKa = 11.84PAGLPATVTSQAVTTRR266 pKa = 11.84PAQQRR271 pKa = 11.84TGRR274 pKa = 11.84HH275 pKa = 5.18LLQNSRR281 pKa = 11.84TGANQGRR288 pKa = 11.84ATAAQGTTSAAIRR301 pKa = 11.84QTVQAANRR309 pKa = 11.84GAPVAAKK316 pKa = 9.19PATRR320 pKa = 11.84TGQAAAQTIARR331 pKa = 11.84PAGLPATVTSQAVTTRR347 pKa = 11.84PAQQNPRR354 pKa = 11.84GLL356 pKa = 3.62
Molecular weight: 35.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10808351 |
11 |
19142 |
583.0 |
61.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
16.312 ± 0.051 | 1.756 ± 0.011 |
4.299 ± 0.012 | 4.558 ± 0.017 |
2.301 ± 0.009 | 8.188 ± 0.02 |
2.148 ± 0.008 | 2.389 ± 0.012 |
3.143 ± 0.016 | 9.543 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.003 ± 0.008 | 2.307 ± 0.012 |
5.784 ± 0.02 | 7.882 ± 0.044 |
5.114 ± 0.013 | 8.719 ± 0.03 |
4.452 ± 0.017 | 6.007 ± 0.016 |
1.271 ± 0.006 | 1.825 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
