
Nocardioides sp. CF8
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
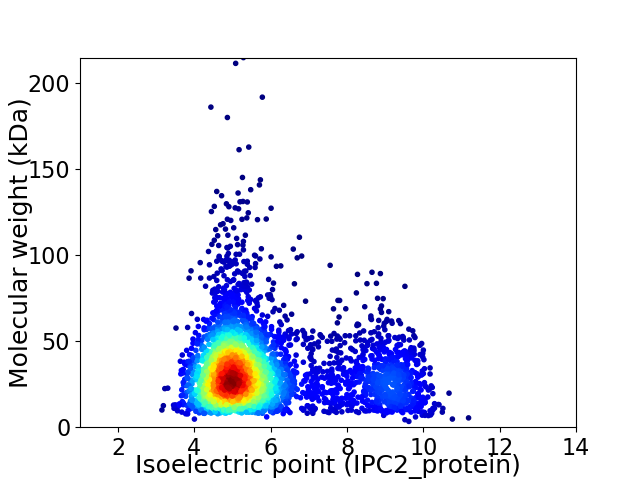
Virtual 2D-PAGE plot for 3893 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
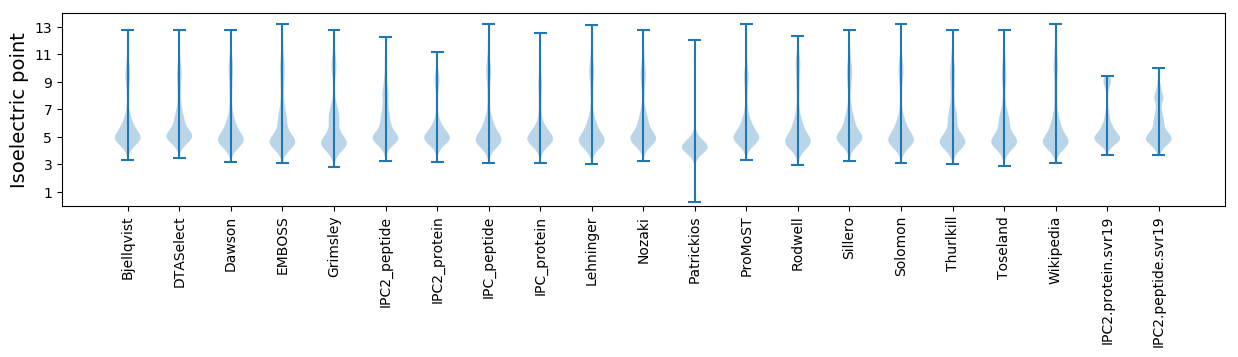
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7XWH7|R7XWH7_9ACTN Uncharacterized protein OS=Nocardioides sp. CF8 OX=110319 GN=CF8_2320 PE=4 SV=1
MM1 pKa = 7.56ALSLHH6 pKa = 5.92SRR8 pKa = 11.84LLITAALVTVAATSGGAAYY27 pKa = 10.34ADD29 pKa = 3.11QVANNLDD36 pKa = 3.61GSVDD40 pKa = 3.31AAAEE44 pKa = 4.06VMPLNVGADD53 pKa = 3.4GSTVLRR59 pKa = 11.84VLTANGDD66 pKa = 3.92GKK68 pKa = 10.55NGCNLTGATIATFSVQSSATDD89 pKa = 3.4VATVTPGTVTFDD101 pKa = 3.1SCEE104 pKa = 3.89RR105 pKa = 11.84TVVLTVTAHH114 pKa = 5.68SVGSSAITLTQTANTTSGSFEE135 pKa = 3.91VDD137 pKa = 2.9PASFTVNVAPPANTAPGVEE156 pKa = 4.12VTGIDD161 pKa = 3.19RR162 pKa = 11.84SSYY165 pKa = 10.83EE166 pKa = 4.3LGLDD170 pKa = 3.47TLPTPGCAATDD181 pKa = 3.89AEE183 pKa = 5.29DD184 pKa = 4.88GPSSPEE190 pKa = 3.61PTVLDD195 pKa = 3.49TRR197 pKa = 11.84DD198 pKa = 3.5ALGLGTVSVSCAYY211 pKa = 9.4TDD213 pKa = 3.43EE214 pKa = 5.39GGLSAVSSQSYY225 pKa = 8.16TVVDD229 pKa = 4.85TIAPVIEE236 pKa = 4.47LVSVTPAAGAHH247 pKa = 5.88GWHH250 pKa = 6.95GSPVTATWSCSDD262 pKa = 3.31AGSGVEE268 pKa = 3.81AAEE271 pKa = 4.39VSATTVGEE279 pKa = 4.32GGDD282 pKa = 4.08LEE284 pKa = 4.79LTDD287 pKa = 3.82TCTDD291 pKa = 3.47RR292 pKa = 11.84AGHH295 pKa = 5.26STSDD299 pKa = 3.24TVEE302 pKa = 4.2DD303 pKa = 4.18LRR305 pKa = 11.84VDD307 pKa = 3.47LTDD310 pKa = 3.54PVITIGRR317 pKa = 11.84SPAANAAGWNNVDD330 pKa = 3.54VTVAWACADD339 pKa = 3.98TLSGTTDD346 pKa = 3.38PGGSAVLGEE355 pKa = 4.65GADD358 pKa = 3.55QSVTATCTDD367 pKa = 3.63LAGNSATDD375 pKa = 3.31EE376 pKa = 4.31VTDD379 pKa = 3.64VDD381 pKa = 3.96VDD383 pKa = 3.64QTAPEE388 pKa = 4.24VSWTSPIADD397 pKa = 3.37GSSYY401 pKa = 11.37YY402 pKa = 10.43FGSVPDD408 pKa = 4.57SPTCSATDD416 pKa = 4.03LLSGLDD422 pKa = 3.66GACAVGGHH430 pKa = 6.02ATTVGTHH437 pKa = 5.1TVTARR442 pKa = 11.84ASDD445 pKa = 3.71VAGNTSQLSSSYY457 pKa = 10.9TVMAWTLNGYY467 pKa = 8.19DD468 pKa = 3.95KK469 pKa = 11.0PVDD472 pKa = 4.0GVGVWNTVKK481 pKa = 10.79NGSTVPLKK489 pKa = 10.84FEE491 pKa = 4.48ALMGSAEE498 pKa = 4.12ITDD501 pKa = 3.87VTTLGAQFTVKK512 pKa = 10.22GVACPGAGAVTEE524 pKa = 4.42DD525 pKa = 4.24VEE527 pKa = 4.47LTTTGGTTFRR537 pKa = 11.84YY538 pKa = 9.44DD539 pKa = 2.75WSGGQFVQNWQTPKK553 pKa = 10.7KK554 pKa = 10.5AGACYY559 pKa = 9.75QVITTTADD567 pKa = 3.33GSSLSALFKK576 pKa = 11.11LKK578 pKa = 10.75
MM1 pKa = 7.56ALSLHH6 pKa = 5.92SRR8 pKa = 11.84LLITAALVTVAATSGGAAYY27 pKa = 10.34ADD29 pKa = 3.11QVANNLDD36 pKa = 3.61GSVDD40 pKa = 3.31AAAEE44 pKa = 4.06VMPLNVGADD53 pKa = 3.4GSTVLRR59 pKa = 11.84VLTANGDD66 pKa = 3.92GKK68 pKa = 10.55NGCNLTGATIATFSVQSSATDD89 pKa = 3.4VATVTPGTVTFDD101 pKa = 3.1SCEE104 pKa = 3.89RR105 pKa = 11.84TVVLTVTAHH114 pKa = 5.68SVGSSAITLTQTANTTSGSFEE135 pKa = 3.91VDD137 pKa = 2.9PASFTVNVAPPANTAPGVEE156 pKa = 4.12VTGIDD161 pKa = 3.19RR162 pKa = 11.84SSYY165 pKa = 10.83EE166 pKa = 4.3LGLDD170 pKa = 3.47TLPTPGCAATDD181 pKa = 3.89AEE183 pKa = 5.29DD184 pKa = 4.88GPSSPEE190 pKa = 3.61PTVLDD195 pKa = 3.49TRR197 pKa = 11.84DD198 pKa = 3.5ALGLGTVSVSCAYY211 pKa = 9.4TDD213 pKa = 3.43EE214 pKa = 5.39GGLSAVSSQSYY225 pKa = 8.16TVVDD229 pKa = 4.85TIAPVIEE236 pKa = 4.47LVSVTPAAGAHH247 pKa = 5.88GWHH250 pKa = 6.95GSPVTATWSCSDD262 pKa = 3.31AGSGVEE268 pKa = 3.81AAEE271 pKa = 4.39VSATTVGEE279 pKa = 4.32GGDD282 pKa = 4.08LEE284 pKa = 4.79LTDD287 pKa = 3.82TCTDD291 pKa = 3.47RR292 pKa = 11.84AGHH295 pKa = 5.26STSDD299 pKa = 3.24TVEE302 pKa = 4.2DD303 pKa = 4.18LRR305 pKa = 11.84VDD307 pKa = 3.47LTDD310 pKa = 3.54PVITIGRR317 pKa = 11.84SPAANAAGWNNVDD330 pKa = 3.54VTVAWACADD339 pKa = 3.98TLSGTTDD346 pKa = 3.38PGGSAVLGEE355 pKa = 4.65GADD358 pKa = 3.55QSVTATCTDD367 pKa = 3.63LAGNSATDD375 pKa = 3.31EE376 pKa = 4.31VTDD379 pKa = 3.64VDD381 pKa = 3.96VDD383 pKa = 3.64QTAPEE388 pKa = 4.24VSWTSPIADD397 pKa = 3.37GSSYY401 pKa = 11.37YY402 pKa = 10.43FGSVPDD408 pKa = 4.57SPTCSATDD416 pKa = 4.03LLSGLDD422 pKa = 3.66GACAVGGHH430 pKa = 6.02ATTVGTHH437 pKa = 5.1TVTARR442 pKa = 11.84ASDD445 pKa = 3.71VAGNTSQLSSSYY457 pKa = 10.9TVMAWTLNGYY467 pKa = 8.19DD468 pKa = 3.95KK469 pKa = 11.0PVDD472 pKa = 4.0GVGVWNTVKK481 pKa = 10.79NGSTVPLKK489 pKa = 10.84FEE491 pKa = 4.48ALMGSAEE498 pKa = 4.12ITDD501 pKa = 3.87VTTLGAQFTVKK512 pKa = 10.22GVACPGAGAVTEE524 pKa = 4.42DD525 pKa = 4.24VEE527 pKa = 4.47LTTTGGTTFRR537 pKa = 11.84YY538 pKa = 9.44DD539 pKa = 2.75WSGGQFVQNWQTPKK553 pKa = 10.7KK554 pKa = 10.5AGACYY559 pKa = 9.75QVITTTADD567 pKa = 3.33GSSLSALFKK576 pKa = 11.11LKK578 pKa = 10.75
Molecular weight: 57.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7XSU3|R7XSU3_9ACTN Integral membrane protein MviN OS=Nocardioides sp. CF8 OX=110319 GN=CF8_3779 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.97KK15 pKa = 10.04VHH17 pKa = 6.59GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.73GRR40 pKa = 11.84SSLAVV45 pKa = 3.16
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.97KK15 pKa = 10.04VHH17 pKa = 6.59GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.73GRR40 pKa = 11.84SSLAVV45 pKa = 3.16
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1249285 |
31 |
1978 |
320.9 |
34.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.802 ± 0.049 | 0.746 ± 0.011 |
6.589 ± 0.031 | 5.844 ± 0.036 |
2.914 ± 0.021 | 8.973 ± 0.035 |
2.291 ± 0.02 | 3.874 ± 0.026 |
2.226 ± 0.034 | 10.185 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.97 ± 0.015 | 1.907 ± 0.022 |
5.388 ± 0.031 | 2.811 ± 0.019 |
7.302 ± 0.04 | 5.482 ± 0.028 |
6.221 ± 0.03 | 9.076 ± 0.037 |
1.499 ± 0.016 | 1.901 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
