
Grapevine leafroll-associated virus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Ampelovirus; unclassified Ampelovirus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
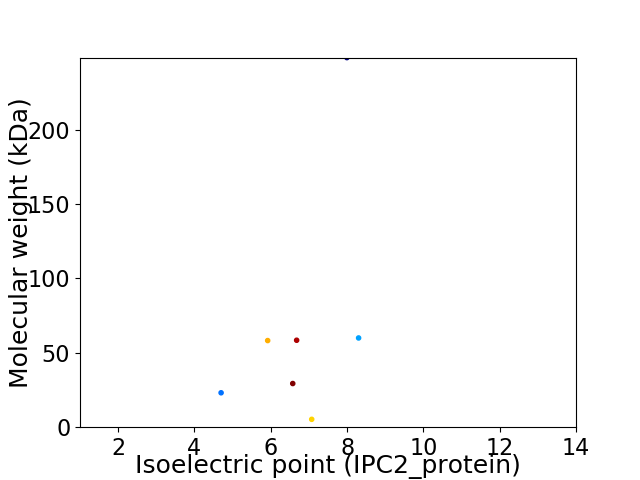
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91U90|Q91U90_9CLOS CPd OS=Grapevine leafroll-associated virus 5 OX=71032 GN=CPm PE=4 SV=1
MM1 pKa = 7.64EE2 pKa = 4.12VTLCLSKK9 pKa = 11.05VKK11 pKa = 10.41EE12 pKa = 4.25KK13 pKa = 11.33LPVTCDD19 pKa = 3.04EE20 pKa = 4.26IFYY23 pKa = 10.66LRR25 pKa = 11.84NAGVYY30 pKa = 8.03ATVMPPEE37 pKa = 4.39RR38 pKa = 11.84LSFFYY43 pKa = 10.95DD44 pKa = 3.19KK45 pKa = 11.41LSVGNNNDD53 pKa = 3.3RR54 pKa = 11.84NKK56 pKa = 10.34YY57 pKa = 9.57RR58 pKa = 11.84EE59 pKa = 4.14LFQFLDD65 pKa = 3.35NLGNVVATNNGNFIKK80 pKa = 10.54FLDD83 pKa = 3.73VDD85 pKa = 3.95SVMEE89 pKa = 4.02YY90 pKa = 10.39FRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84YY95 pKa = 9.2PDD97 pKa = 3.37GGNLSSINKK106 pKa = 8.37TPSSDD111 pKa = 3.95YY112 pKa = 11.27CCLTVPALTTFLNSVCGCIEE132 pKa = 3.94GLTTVLAVGFTRR144 pKa = 11.84KK145 pKa = 9.74SDD147 pKa = 3.61TDD149 pKa = 3.74EE150 pKa = 3.93LWSFKK155 pKa = 9.71NTFVVFEE162 pKa = 4.12NSGEE166 pKa = 3.84ISTFYY171 pKa = 10.37GDD173 pKa = 5.9SMDD176 pKa = 3.76TSNSALLEE184 pKa = 4.12VLDD187 pKa = 4.66SFGSTSGKK195 pKa = 8.61TDD197 pKa = 3.58FEE199 pKa = 4.65KK200 pKa = 11.03VVSLLSII207 pKa = 4.28
MM1 pKa = 7.64EE2 pKa = 4.12VTLCLSKK9 pKa = 11.05VKK11 pKa = 10.41EE12 pKa = 4.25KK13 pKa = 11.33LPVTCDD19 pKa = 3.04EE20 pKa = 4.26IFYY23 pKa = 10.66LRR25 pKa = 11.84NAGVYY30 pKa = 8.03ATVMPPEE37 pKa = 4.39RR38 pKa = 11.84LSFFYY43 pKa = 10.95DD44 pKa = 3.19KK45 pKa = 11.41LSVGNNNDD53 pKa = 3.3RR54 pKa = 11.84NKK56 pKa = 10.34YY57 pKa = 9.57RR58 pKa = 11.84EE59 pKa = 4.14LFQFLDD65 pKa = 3.35NLGNVVATNNGNFIKK80 pKa = 10.54FLDD83 pKa = 3.73VDD85 pKa = 3.95SVMEE89 pKa = 4.02YY90 pKa = 10.39FRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84YY95 pKa = 9.2PDD97 pKa = 3.37GGNLSSINKK106 pKa = 8.37TPSSDD111 pKa = 3.95YY112 pKa = 11.27CCLTVPALTTFLNSVCGCIEE132 pKa = 3.94GLTTVLAVGFTRR144 pKa = 11.84KK145 pKa = 9.74SDD147 pKa = 3.61TDD149 pKa = 3.74EE150 pKa = 3.93LWSFKK155 pKa = 9.71NTFVVFEE162 pKa = 4.12NSGEE166 pKa = 3.84ISTFYY171 pKa = 10.37GDD173 pKa = 5.9SMDD176 pKa = 3.76TSNSALLEE184 pKa = 4.12VLDD187 pKa = 4.66SFGSTSGKK195 pKa = 8.61TDD197 pKa = 3.58FEE199 pKa = 4.65KK200 pKa = 11.03VVSLLSII207 pKa = 4.28
Molecular weight: 23.13 kDa
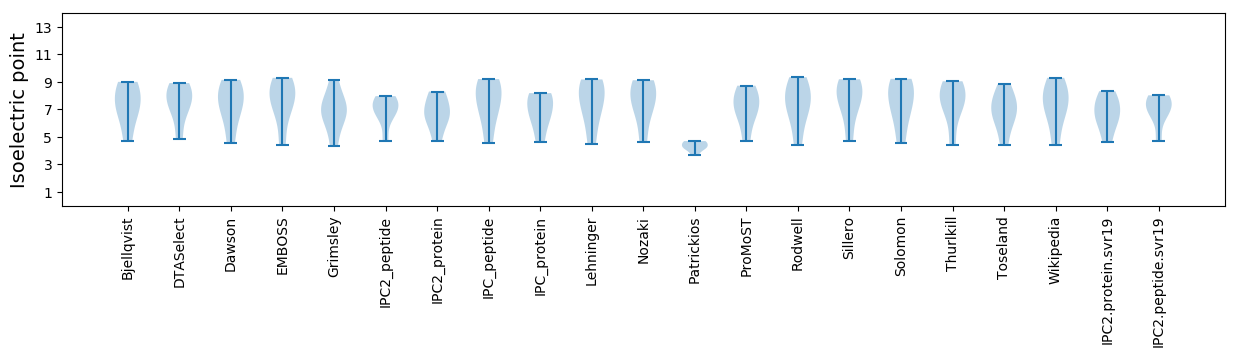
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G4U4B0|G4U4B0_9CLOS Coat protein OS=Grapevine leafroll-associated virus 5 OX=71032 GN=CP PE=4 SV=1
MM1 pKa = 7.53ALSATSTYY9 pKa = 10.67VRR11 pKa = 11.84VSKK14 pKa = 10.8SDD16 pKa = 3.31EE17 pKa = 4.07GFVNLLRR24 pKa = 11.84TFKK27 pKa = 11.28GEE29 pKa = 3.91NDD31 pKa = 2.74VDD33 pKa = 3.8AFADD37 pKa = 3.8EE38 pKa = 4.25VLNYY42 pKa = 10.64LDD44 pKa = 5.07LRR46 pKa = 11.84FSAISGRR53 pKa = 11.84FSLGNYY59 pKa = 9.2SCNTWYY65 pKa = 10.89NGVSGAVGAWPDD77 pKa = 3.37SEE79 pKa = 3.89GWGRR83 pKa = 11.84HH84 pKa = 4.73LLANYY89 pKa = 9.24IVSNNALAHH98 pKa = 6.63LYY100 pKa = 10.36YY101 pKa = 10.31PLASEE106 pKa = 4.46IKK108 pKa = 9.82SAVATPMSVFTDD120 pKa = 3.1KK121 pKa = 10.95LSRR124 pKa = 11.84VGKK127 pKa = 9.29PVTEE131 pKa = 3.98TSKK134 pKa = 10.98FPFKK138 pKa = 10.61VSRR141 pKa = 11.84VEE143 pKa = 4.06AEE145 pKa = 3.85KK146 pKa = 10.77KK147 pKa = 9.84AAEE150 pKa = 4.06FGKK153 pKa = 8.31PTKK156 pKa = 9.34HH157 pKa = 5.12TQDD160 pKa = 4.61LIFCIGNYY168 pKa = 9.92LGRR171 pKa = 11.84LPTKK175 pKa = 10.4DD176 pKa = 3.44EE177 pKa = 4.22VMGMTSLPVAAVRR190 pKa = 11.84KK191 pKa = 9.91GSLLLDD197 pKa = 3.63VGSDD201 pKa = 3.33VTSNEE206 pKa = 3.86RR207 pKa = 11.84KK208 pKa = 9.64LATLNLRR215 pKa = 11.84GKK217 pKa = 9.53NKK219 pKa = 10.18QGDD222 pKa = 3.78PVEE225 pKa = 4.47GVFFKK230 pKa = 10.51TRR232 pKa = 11.84QTEE235 pKa = 4.07IEE237 pKa = 4.38TVASFIFDD245 pKa = 3.37NKK247 pKa = 9.6IVKK250 pKa = 7.72EE251 pKa = 4.18TLSLPGVQWTILSCLKK267 pKa = 10.17KK268 pKa = 10.61YY269 pKa = 10.41PVFADD274 pKa = 3.51SFEE277 pKa = 4.36DD278 pKa = 4.47RR279 pKa = 11.84IDD281 pKa = 3.54SLVYY285 pKa = 10.13VQAQVRR291 pKa = 11.84KK292 pKa = 9.29LLDD295 pKa = 3.23GLIPFKK301 pKa = 11.04EE302 pKa = 5.06DD303 pKa = 2.88IDD305 pKa = 4.64LDD307 pKa = 3.98AALSSGTLLKK317 pKa = 11.26GEE319 pKa = 4.42VFRR322 pKa = 11.84GDD324 pKa = 3.5AKK326 pKa = 11.15VLTPRR331 pKa = 11.84FQRR334 pKa = 11.84NTTGGLLMRR343 pKa = 11.84IGSRR347 pKa = 11.84FSVSEE352 pKa = 3.86YY353 pKa = 11.12SEE355 pKa = 4.12LTRR358 pKa = 11.84KK359 pKa = 9.7LVGALLNRR367 pKa = 11.84NLSMDD372 pKa = 3.39VTYY375 pKa = 9.2TQAVLILLQRR385 pKa = 11.84YY386 pKa = 6.9IHH388 pKa = 6.1YY389 pKa = 7.76RR390 pKa = 11.84TNALRR395 pKa = 11.84YY396 pKa = 8.91VNLPAKK402 pKa = 10.01LDD404 pKa = 3.61YY405 pKa = 10.2MIKK408 pKa = 10.54NEE410 pKa = 3.92VKK412 pKa = 9.98QVDD415 pKa = 4.09FSGVDD420 pKa = 3.52EE421 pKa = 4.78VFSKK425 pKa = 11.13YY426 pKa = 10.07QGTIPEE432 pKa = 4.81IEE434 pKa = 4.44RR435 pKa = 11.84AWCAPLADD443 pKa = 3.75VAYY446 pKa = 9.66MFLKK450 pKa = 10.12DD451 pKa = 3.55TGGSFAKK458 pKa = 9.92WKK460 pKa = 10.32DD461 pKa = 3.72VQDD464 pKa = 4.21VPAHH468 pKa = 5.63MNFDD472 pKa = 3.65FVGFVDD478 pKa = 4.82PKK480 pKa = 10.81LLSDD484 pKa = 5.6HH485 pKa = 6.25EE486 pKa = 4.89LKK488 pKa = 10.98CQTRR492 pKa = 11.84LLDD495 pKa = 3.53RR496 pKa = 11.84FRR498 pKa = 11.84SKK500 pKa = 10.06DD501 pKa = 3.31TPVRR505 pKa = 11.84GFLLGARR512 pKa = 11.84KK513 pKa = 9.77GNPVDD518 pKa = 4.01YY519 pKa = 9.62LASSAGIGGIEE530 pKa = 3.77KK531 pKa = 10.91AMVKK535 pKa = 10.41KK536 pKa = 10.96LIGG539 pKa = 3.55
MM1 pKa = 7.53ALSATSTYY9 pKa = 10.67VRR11 pKa = 11.84VSKK14 pKa = 10.8SDD16 pKa = 3.31EE17 pKa = 4.07GFVNLLRR24 pKa = 11.84TFKK27 pKa = 11.28GEE29 pKa = 3.91NDD31 pKa = 2.74VDD33 pKa = 3.8AFADD37 pKa = 3.8EE38 pKa = 4.25VLNYY42 pKa = 10.64LDD44 pKa = 5.07LRR46 pKa = 11.84FSAISGRR53 pKa = 11.84FSLGNYY59 pKa = 9.2SCNTWYY65 pKa = 10.89NGVSGAVGAWPDD77 pKa = 3.37SEE79 pKa = 3.89GWGRR83 pKa = 11.84HH84 pKa = 4.73LLANYY89 pKa = 9.24IVSNNALAHH98 pKa = 6.63LYY100 pKa = 10.36YY101 pKa = 10.31PLASEE106 pKa = 4.46IKK108 pKa = 9.82SAVATPMSVFTDD120 pKa = 3.1KK121 pKa = 10.95LSRR124 pKa = 11.84VGKK127 pKa = 9.29PVTEE131 pKa = 3.98TSKK134 pKa = 10.98FPFKK138 pKa = 10.61VSRR141 pKa = 11.84VEE143 pKa = 4.06AEE145 pKa = 3.85KK146 pKa = 10.77KK147 pKa = 9.84AAEE150 pKa = 4.06FGKK153 pKa = 8.31PTKK156 pKa = 9.34HH157 pKa = 5.12TQDD160 pKa = 4.61LIFCIGNYY168 pKa = 9.92LGRR171 pKa = 11.84LPTKK175 pKa = 10.4DD176 pKa = 3.44EE177 pKa = 4.22VMGMTSLPVAAVRR190 pKa = 11.84KK191 pKa = 9.91GSLLLDD197 pKa = 3.63VGSDD201 pKa = 3.33VTSNEE206 pKa = 3.86RR207 pKa = 11.84KK208 pKa = 9.64LATLNLRR215 pKa = 11.84GKK217 pKa = 9.53NKK219 pKa = 10.18QGDD222 pKa = 3.78PVEE225 pKa = 4.47GVFFKK230 pKa = 10.51TRR232 pKa = 11.84QTEE235 pKa = 4.07IEE237 pKa = 4.38TVASFIFDD245 pKa = 3.37NKK247 pKa = 9.6IVKK250 pKa = 7.72EE251 pKa = 4.18TLSLPGVQWTILSCLKK267 pKa = 10.17KK268 pKa = 10.61YY269 pKa = 10.41PVFADD274 pKa = 3.51SFEE277 pKa = 4.36DD278 pKa = 4.47RR279 pKa = 11.84IDD281 pKa = 3.54SLVYY285 pKa = 10.13VQAQVRR291 pKa = 11.84KK292 pKa = 9.29LLDD295 pKa = 3.23GLIPFKK301 pKa = 11.04EE302 pKa = 5.06DD303 pKa = 2.88IDD305 pKa = 4.64LDD307 pKa = 3.98AALSSGTLLKK317 pKa = 11.26GEE319 pKa = 4.42VFRR322 pKa = 11.84GDD324 pKa = 3.5AKK326 pKa = 11.15VLTPRR331 pKa = 11.84FQRR334 pKa = 11.84NTTGGLLMRR343 pKa = 11.84IGSRR347 pKa = 11.84FSVSEE352 pKa = 3.86YY353 pKa = 11.12SEE355 pKa = 4.12LTRR358 pKa = 11.84KK359 pKa = 9.7LVGALLNRR367 pKa = 11.84NLSMDD372 pKa = 3.39VTYY375 pKa = 9.2TQAVLILLQRR385 pKa = 11.84YY386 pKa = 6.9IHH388 pKa = 6.1YY389 pKa = 7.76RR390 pKa = 11.84TNALRR395 pKa = 11.84YY396 pKa = 8.91VNLPAKK402 pKa = 10.01LDD404 pKa = 3.61YY405 pKa = 10.2MIKK408 pKa = 10.54NEE410 pKa = 3.92VKK412 pKa = 9.98QVDD415 pKa = 4.09FSGVDD420 pKa = 3.52EE421 pKa = 4.78VFSKK425 pKa = 11.13YY426 pKa = 10.07QGTIPEE432 pKa = 4.81IEE434 pKa = 4.44RR435 pKa = 11.84AWCAPLADD443 pKa = 3.75VAYY446 pKa = 9.66MFLKK450 pKa = 10.12DD451 pKa = 3.55TGGSFAKK458 pKa = 9.92WKK460 pKa = 10.32DD461 pKa = 3.72VQDD464 pKa = 4.21VPAHH468 pKa = 5.63MNFDD472 pKa = 3.65FVGFVDD478 pKa = 4.82PKK480 pKa = 10.81LLSDD484 pKa = 5.6HH485 pKa = 6.25EE486 pKa = 4.89LKK488 pKa = 10.98CQTRR492 pKa = 11.84LLDD495 pKa = 3.53RR496 pKa = 11.84FRR498 pKa = 11.84SKK500 pKa = 10.06DD501 pKa = 3.31TPVRR505 pKa = 11.84GFLLGARR512 pKa = 11.84KK513 pKa = 9.77GNPVDD518 pKa = 4.01YY519 pKa = 9.62LASSAGIGGIEE530 pKa = 3.77KK531 pKa = 10.91AMVKK535 pKa = 10.41KK536 pKa = 10.96LIGG539 pKa = 3.55
Molecular weight: 59.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4352 |
46 |
2241 |
621.7 |
68.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.457 ± 0.308 | 1.93 ± 0.248 |
5.561 ± 0.496 | 5.699 ± 0.319 |
5.17 ± 0.817 | 6.043 ± 0.462 |
1.517 ± 0.381 | 5.055 ± 0.366 |
7.146 ± 0.594 | 9.099 ± 0.427 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.091 ± 0.178 | 5.446 ± 0.284 |
4.251 ± 0.278 | 2.229 ± 0.182 |
4.986 ± 0.16 | 9.513 ± 0.577 |
5.767 ± 0.288 | 8.456 ± 0.59 |
0.735 ± 0.144 | 2.849 ± 0.356 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
