
Elysia chlorotica (Eastern emerald elysia) (Sea slug)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Mollusca; Gastropoda; Heterobranchia; Euthyneura; Panpulmonata; Sacoglossa; Placobranchoidea; Plakobranchidae; Elysia
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
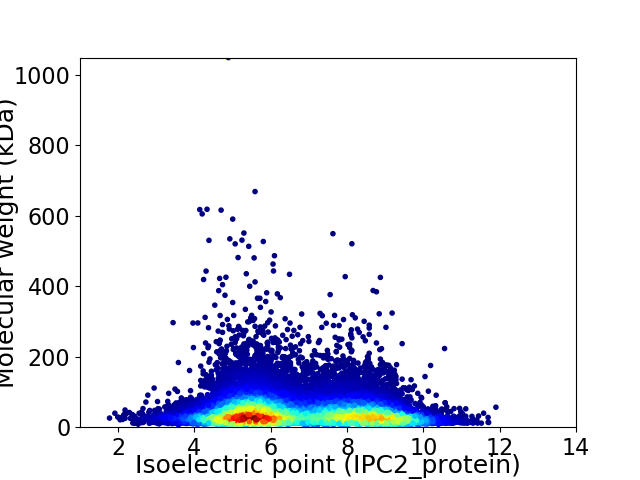
Virtual 2D-PAGE plot for 23836 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S1I3S8|A0A3S1I3S8_ELYCH Uncharacterized protein OS=Elysia chlorotica OX=188477 GN=EGW08_000503 PE=4 SV=1
MM1 pKa = 7.4KK2 pKa = 9.73PAVNPVSTINKK13 pKa = 6.69TLPGVTGHH21 pKa = 5.8VHH23 pKa = 5.79HH24 pKa = 7.01HH25 pKa = 6.09WSCGGTIVSISHH37 pKa = 7.07PLGCLSSFTVEE48 pKa = 4.33MKK50 pKa = 10.31TGMSDD55 pKa = 4.36DD56 pKa = 4.24GACGGDD62 pKa = 3.1GGGGFKK68 pKa = 11.0DD69 pKa = 3.55EE70 pKa = 5.6DD71 pKa = 3.92EE72 pKa = 4.43YY73 pKa = 11.55DD74 pKa = 3.67YY75 pKa = 11.68DD76 pKa = 3.61SGGGCRR82 pKa = 11.84VGGVGDD88 pKa = 4.29CGVSGGGGCGFGGGGGFGLGGGGGFGVGGVGVCGVGGVGGVDD130 pKa = 3.69YY131 pKa = 11.43GNDD134 pKa = 3.73AADD137 pKa = 4.56DD138 pKa = 5.11GYY140 pKa = 11.53DD141 pKa = 3.28SAAADD146 pKa = 4.6GDD148 pKa = 3.96DD149 pKa = 4.44DD150 pKa = 4.79QNGDD154 pKa = 3.33GDD156 pKa = 4.36YY157 pKa = 10.8EE158 pKa = 4.07AAAAAAAAAADD169 pKa = 5.07DD170 pKa = 5.87DD171 pKa = 6.36DD172 pKa = 7.71DD173 pKa = 7.66DD174 pKa = 7.66DD175 pKa = 7.66DD176 pKa = 7.65DD177 pKa = 7.65DD178 pKa = 7.65DD179 pKa = 7.65DD180 pKa = 7.65DD181 pKa = 7.65DD182 pKa = 7.65DD183 pKa = 7.65DD184 pKa = 7.65DD185 pKa = 7.65DD186 pKa = 7.61DD187 pKa = 7.6DD188 pKa = 7.65DD189 pKa = 7.6DD190 pKa = 5.71DD191 pKa = 5.5DD192 pKa = 6.05CGGVDD197 pKa = 4.32DD198 pKa = 4.49MMIVVSRR205 pKa = 11.84RR206 pKa = 11.84MVQIAPQLPGRR217 pKa = 11.84LKK219 pKa = 10.86KK220 pKa = 10.65EE221 pKa = 4.27GGSGPTTKK229 pKa = 10.29AYY231 pKa = 8.29PQTRR235 pKa = 11.84VLYY238 pKa = 10.96AEE240 pKa = 5.31VNDD243 pKa = 3.81QCGEE247 pKa = 3.77RR248 pKa = 11.84STNRR252 pKa = 11.84FCGSLRR258 pKa = 11.84TFRR261 pKa = 11.84LIEE264 pKa = 4.26PKK266 pKa = 9.51WNPISQPRR274 pKa = 11.84YY275 pKa = 7.23TVLWWSPNGVQPQTPCLSPPTSMSHH300 pKa = 5.3YY301 pKa = 10.11AYY303 pKa = 9.63IDD305 pKa = 3.27ATSRR309 pKa = 11.84EE310 pKa = 4.11QGDD313 pKa = 4.01PLWIGTQGFLTSFSGLDD330 pKa = 3.33MLVLSVSEE338 pKa = 3.96WLGLGVGGG346 pKa = 4.95
MM1 pKa = 7.4KK2 pKa = 9.73PAVNPVSTINKK13 pKa = 6.69TLPGVTGHH21 pKa = 5.8VHH23 pKa = 5.79HH24 pKa = 7.01HH25 pKa = 6.09WSCGGTIVSISHH37 pKa = 7.07PLGCLSSFTVEE48 pKa = 4.33MKK50 pKa = 10.31TGMSDD55 pKa = 4.36DD56 pKa = 4.24GACGGDD62 pKa = 3.1GGGGFKK68 pKa = 11.0DD69 pKa = 3.55EE70 pKa = 5.6DD71 pKa = 3.92EE72 pKa = 4.43YY73 pKa = 11.55DD74 pKa = 3.67YY75 pKa = 11.68DD76 pKa = 3.61SGGGCRR82 pKa = 11.84VGGVGDD88 pKa = 4.29CGVSGGGGCGFGGGGGFGLGGGGGFGVGGVGVCGVGGVGGVDD130 pKa = 3.69YY131 pKa = 11.43GNDD134 pKa = 3.73AADD137 pKa = 4.56DD138 pKa = 5.11GYY140 pKa = 11.53DD141 pKa = 3.28SAAADD146 pKa = 4.6GDD148 pKa = 3.96DD149 pKa = 4.44DD150 pKa = 4.79QNGDD154 pKa = 3.33GDD156 pKa = 4.36YY157 pKa = 10.8EE158 pKa = 4.07AAAAAAAAAADD169 pKa = 5.07DD170 pKa = 5.87DD171 pKa = 6.36DD172 pKa = 7.71DD173 pKa = 7.66DD174 pKa = 7.66DD175 pKa = 7.66DD176 pKa = 7.65DD177 pKa = 7.65DD178 pKa = 7.65DD179 pKa = 7.65DD180 pKa = 7.65DD181 pKa = 7.65DD182 pKa = 7.65DD183 pKa = 7.65DD184 pKa = 7.65DD185 pKa = 7.65DD186 pKa = 7.61DD187 pKa = 7.6DD188 pKa = 7.65DD189 pKa = 7.6DD190 pKa = 5.71DD191 pKa = 5.5DD192 pKa = 6.05CGGVDD197 pKa = 4.32DD198 pKa = 4.49MMIVVSRR205 pKa = 11.84RR206 pKa = 11.84MVQIAPQLPGRR217 pKa = 11.84LKK219 pKa = 10.86KK220 pKa = 10.65EE221 pKa = 4.27GGSGPTTKK229 pKa = 10.29AYY231 pKa = 8.29PQTRR235 pKa = 11.84VLYY238 pKa = 10.96AEE240 pKa = 5.31VNDD243 pKa = 3.81QCGEE247 pKa = 3.77RR248 pKa = 11.84STNRR252 pKa = 11.84FCGSLRR258 pKa = 11.84TFRR261 pKa = 11.84LIEE264 pKa = 4.26PKK266 pKa = 9.51WNPISQPRR274 pKa = 11.84YY275 pKa = 7.23TVLWWSPNGVQPQTPCLSPPTSMSHH300 pKa = 5.3YY301 pKa = 10.11AYY303 pKa = 9.63IDD305 pKa = 3.27ATSRR309 pKa = 11.84EE310 pKa = 4.11QGDD313 pKa = 4.01PLWIGTQGFLTSFSGLDD330 pKa = 3.33MLVLSVSEE338 pKa = 3.96WLGLGVGGG346 pKa = 4.95
Molecular weight: 35.45 kDa
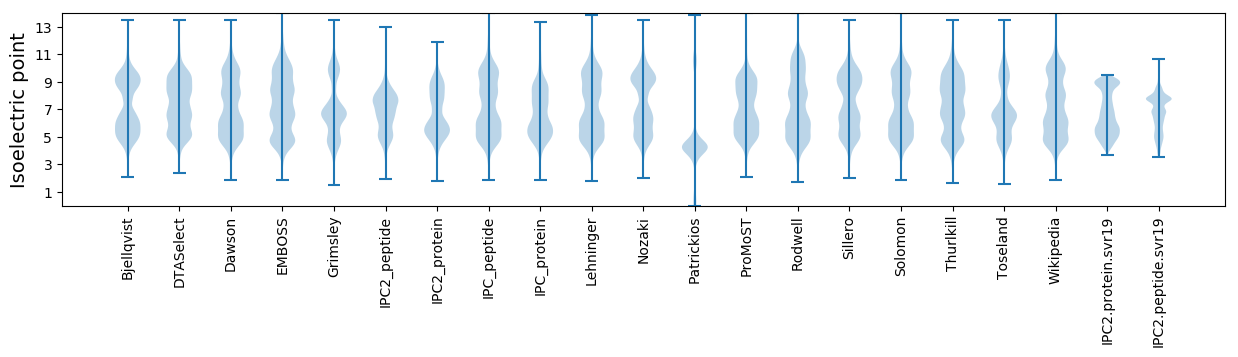
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A433U0U8|A0A433U0U8_ELYCH Uncharacterized protein OS=Elysia chlorotica OX=188477 GN=EGW08_004785 PE=4 SV=1
MM1 pKa = 7.08TPLVARR7 pKa = 11.84NIPPLVARR15 pKa = 11.84NILPLVARR23 pKa = 11.84NILPLVARR31 pKa = 11.84NIPPLVARR39 pKa = 11.84NILPLVAGNIQLLVARR55 pKa = 11.84HH56 pKa = 5.98ILPLVARR63 pKa = 11.84NILPLVARR71 pKa = 11.84NILPLVARR79 pKa = 11.84NILPLVARR87 pKa = 11.84NILPLVARR95 pKa = 11.84NIIRR99 pKa = 11.84LFARR103 pKa = 11.84HH104 pKa = 5.35NN105 pKa = 3.55
MM1 pKa = 7.08TPLVARR7 pKa = 11.84NIPPLVARR15 pKa = 11.84NILPLVARR23 pKa = 11.84NILPLVARR31 pKa = 11.84NIPPLVARR39 pKa = 11.84NILPLVAGNIQLLVARR55 pKa = 11.84HH56 pKa = 5.98ILPLVARR63 pKa = 11.84NILPLVARR71 pKa = 11.84NILPLVARR79 pKa = 11.84NILPLVARR87 pKa = 11.84NILPLVARR95 pKa = 11.84NIIRR99 pKa = 11.84LFARR103 pKa = 11.84HH104 pKa = 5.35NN105 pKa = 3.55
Molecular weight: 11.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10343547 |
50 |
9454 |
433.9 |
48.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.625 ± 0.014 | 2.11 ± 0.017 |
5.912 ± 0.02 | 6.198 ± 0.024 |
3.659 ± 0.013 | 6.296 ± 0.018 |
2.685 ± 0.01 | 4.567 ± 0.013 |
5.817 ± 0.02 | 8.777 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.364 ± 0.009 | 4.324 ± 0.013 |
5.288 ± 0.019 | 4.474 ± 0.016 |
5.617 ± 0.017 | 9.165 ± 0.029 |
5.897 ± 0.016 | 6.335 ± 0.018 |
1.087 ± 0.007 | 2.772 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
