
Microbacterium testaceum (strain StLB037)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; Microbacterium testaceum
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
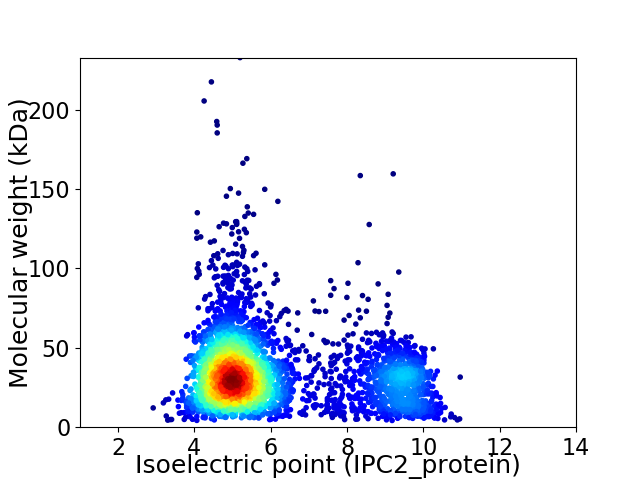
Virtual 2D-PAGE plot for 3671 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
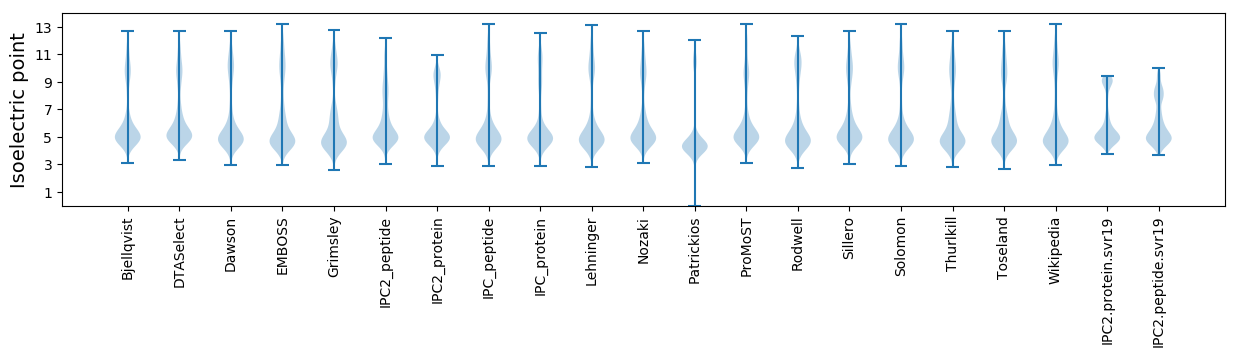
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E8N7F3|E8N7F3_MICTS Predicted ferric reductase OS=Microbacterium testaceum (strain StLB037) OX=979556 GN=MTES_0022 PE=4 SV=1
MM1 pKa = 7.32SRR3 pKa = 11.84IPDD6 pKa = 3.56TQMTPEE12 pKa = 4.45GPAYY16 pKa = 9.92EE17 pKa = 4.59GRR19 pKa = 11.84LLDD22 pKa = 4.08RR23 pKa = 11.84ADD25 pKa = 4.12EE26 pKa = 4.22PVVDD30 pKa = 4.31QGAPFDD36 pKa = 3.53IRR38 pKa = 11.84TLVSRR43 pKa = 11.84RR44 pKa = 11.84GILGLVALGAGAVTLVACTPAATGTTTATTATPTATASATPTASATTSAAALPAGEE100 pKa = 5.09IPDD103 pKa = 4.05EE104 pKa = 4.33TAGPYY109 pKa = 10.2PGDD112 pKa = 3.75GSNGADD118 pKa = 3.21VLEE121 pKa = 4.02QSGIVRR127 pKa = 11.84SDD129 pKa = 3.15LRR131 pKa = 11.84SSLDD135 pKa = 3.27GGATAAGTPMTLNLTIFDD153 pKa = 4.28TANGDD158 pKa = 3.74TPMTGAAVYY167 pKa = 9.65VWHH170 pKa = 7.65CDD172 pKa = 3.29TEE174 pKa = 5.0GNYY177 pKa = 11.2SMYY180 pKa = 10.83SSGIEE185 pKa = 3.74NEE187 pKa = 4.14TYY189 pKa = 10.54LRR191 pKa = 11.84GVQVVGDD198 pKa = 4.12DD199 pKa = 3.87GVVSFTSIFPGCYY212 pKa = 9.22DD213 pKa = 3.33GRR215 pKa = 11.84WPHH218 pKa = 6.03IHH220 pKa = 6.34FEE222 pKa = 4.43VYY224 pKa = 10.23PSVDD228 pKa = 3.97AITDD232 pKa = 3.54ASNAIATSQVALPKK246 pKa = 8.18EE247 pKa = 4.5TCDD250 pKa = 3.44VVYY253 pKa = 10.73ADD255 pKa = 3.58TSLYY259 pKa = 9.92PSSASNLTRR268 pKa = 11.84VSLDD272 pKa = 3.04SDD274 pKa = 3.81NVFGDD279 pKa = 4.32DD280 pKa = 4.91GGALQLATTSGDD292 pKa = 3.31NASGYY297 pKa = 7.7TVSLGVRR304 pKa = 11.84VDD306 pKa = 3.45TATTPTAGAAPGGRR320 pKa = 3.66
MM1 pKa = 7.32SRR3 pKa = 11.84IPDD6 pKa = 3.56TQMTPEE12 pKa = 4.45GPAYY16 pKa = 9.92EE17 pKa = 4.59GRR19 pKa = 11.84LLDD22 pKa = 4.08RR23 pKa = 11.84ADD25 pKa = 4.12EE26 pKa = 4.22PVVDD30 pKa = 4.31QGAPFDD36 pKa = 3.53IRR38 pKa = 11.84TLVSRR43 pKa = 11.84RR44 pKa = 11.84GILGLVALGAGAVTLVACTPAATGTTTATTATPTATASATPTASATTSAAALPAGEE100 pKa = 5.09IPDD103 pKa = 4.05EE104 pKa = 4.33TAGPYY109 pKa = 10.2PGDD112 pKa = 3.75GSNGADD118 pKa = 3.21VLEE121 pKa = 4.02QSGIVRR127 pKa = 11.84SDD129 pKa = 3.15LRR131 pKa = 11.84SSLDD135 pKa = 3.27GGATAAGTPMTLNLTIFDD153 pKa = 4.28TANGDD158 pKa = 3.74TPMTGAAVYY167 pKa = 9.65VWHH170 pKa = 7.65CDD172 pKa = 3.29TEE174 pKa = 5.0GNYY177 pKa = 11.2SMYY180 pKa = 10.83SSGIEE185 pKa = 3.74NEE187 pKa = 4.14TYY189 pKa = 10.54LRR191 pKa = 11.84GVQVVGDD198 pKa = 4.12DD199 pKa = 3.87GVVSFTSIFPGCYY212 pKa = 9.22DD213 pKa = 3.33GRR215 pKa = 11.84WPHH218 pKa = 6.03IHH220 pKa = 6.34FEE222 pKa = 4.43VYY224 pKa = 10.23PSVDD228 pKa = 3.97AITDD232 pKa = 3.54ASNAIATSQVALPKK246 pKa = 8.18EE247 pKa = 4.5TCDD250 pKa = 3.44VVYY253 pKa = 10.73ADD255 pKa = 3.58TSLYY259 pKa = 9.92PSSASNLTRR268 pKa = 11.84VSLDD272 pKa = 3.04SDD274 pKa = 3.81NVFGDD279 pKa = 4.32DD280 pKa = 4.91GGALQLATTSGDD292 pKa = 3.31NASGYY297 pKa = 7.7TVSLGVRR304 pKa = 11.84VDD306 pKa = 3.45TATTPTAGAAPGGRR320 pKa = 3.66
Molecular weight: 32.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E8NER9|E8NER9_MICTS D-inositol 3-phosphate glycosyltransferase OS=Microbacterium testaceum (strain StLB037) OX=979556 GN=MTES_2190 PE=4 SV=1
MM1 pKa = 6.81GTVPRR6 pKa = 11.84AATGIARR13 pKa = 11.84SVATMMRR20 pKa = 11.84VRR22 pKa = 11.84RR23 pKa = 11.84AMATVRR29 pKa = 11.84SVATTTGVPLEE40 pKa = 4.07MAIVLSVVMVRR51 pKa = 11.84ARR53 pKa = 11.84GVPRR57 pKa = 11.84VMATVLSVATTMRR70 pKa = 11.84VRR72 pKa = 11.84RR73 pKa = 11.84AMATVLSVGTTMRR86 pKa = 11.84GPRR89 pKa = 11.84GMAIVLSVATTMRR102 pKa = 11.84VPRR105 pKa = 11.84VMATVLSAGTTMRR118 pKa = 11.84VRR120 pKa = 11.84RR121 pKa = 11.84GMAIVLSVGTTMRR134 pKa = 11.84GPRR137 pKa = 11.84GMAIVLSVGTTMRR150 pKa = 11.84GPRR153 pKa = 11.84GMAIVLSVATTTGVRR168 pKa = 11.84RR169 pKa = 11.84GTVIVLSVVTASVRR183 pKa = 11.84SDD185 pKa = 3.03ATVMDD190 pKa = 3.63VRR192 pKa = 11.84RR193 pKa = 11.84ARR195 pKa = 11.84VTVGRR200 pKa = 11.84NATAIVLSTVTLVAPVDD217 pKa = 4.08RR218 pKa = 11.84PVMATVHH225 pKa = 5.84SAVTATPPGARR236 pKa = 11.84GVTVSRR242 pKa = 11.84AASAGVTVAPPATVGPVPAHH262 pKa = 6.17LAVSTVRR269 pKa = 11.84TPARR273 pKa = 11.84RR274 pKa = 11.84DD275 pKa = 3.4RR276 pKa = 11.84LRR278 pKa = 11.84IAANAARR285 pKa = 11.84RR286 pKa = 11.84FRR288 pKa = 11.84KK289 pKa = 9.37RR290 pKa = 11.84SPRR293 pKa = 11.84GTCRR297 pKa = 11.84ALRR300 pKa = 11.84AMNN303 pKa = 4.05
MM1 pKa = 6.81GTVPRR6 pKa = 11.84AATGIARR13 pKa = 11.84SVATMMRR20 pKa = 11.84VRR22 pKa = 11.84RR23 pKa = 11.84AMATVRR29 pKa = 11.84SVATTTGVPLEE40 pKa = 4.07MAIVLSVVMVRR51 pKa = 11.84ARR53 pKa = 11.84GVPRR57 pKa = 11.84VMATVLSVATTMRR70 pKa = 11.84VRR72 pKa = 11.84RR73 pKa = 11.84AMATVLSVGTTMRR86 pKa = 11.84GPRR89 pKa = 11.84GMAIVLSVATTMRR102 pKa = 11.84VPRR105 pKa = 11.84VMATVLSAGTTMRR118 pKa = 11.84VRR120 pKa = 11.84RR121 pKa = 11.84GMAIVLSVGTTMRR134 pKa = 11.84GPRR137 pKa = 11.84GMAIVLSVGTTMRR150 pKa = 11.84GPRR153 pKa = 11.84GMAIVLSVATTTGVRR168 pKa = 11.84RR169 pKa = 11.84GTVIVLSVVTASVRR183 pKa = 11.84SDD185 pKa = 3.03ATVMDD190 pKa = 3.63VRR192 pKa = 11.84RR193 pKa = 11.84ARR195 pKa = 11.84VTVGRR200 pKa = 11.84NATAIVLSTVTLVAPVDD217 pKa = 4.08RR218 pKa = 11.84PVMATVHH225 pKa = 5.84SAVTATPPGARR236 pKa = 11.84GVTVSRR242 pKa = 11.84AASAGVTVAPPATVGPVPAHH262 pKa = 6.17LAVSTVRR269 pKa = 11.84TPARR273 pKa = 11.84RR274 pKa = 11.84DD275 pKa = 3.4RR276 pKa = 11.84LRR278 pKa = 11.84IAANAARR285 pKa = 11.84RR286 pKa = 11.84FRR288 pKa = 11.84KK289 pKa = 9.37RR290 pKa = 11.84SPRR293 pKa = 11.84GTCRR297 pKa = 11.84ALRR300 pKa = 11.84AMNN303 pKa = 4.05
Molecular weight: 31.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1212999 |
39 |
2162 |
330.4 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.925 ± 0.063 | 0.465 ± 0.008 |
6.435 ± 0.04 | 5.381 ± 0.042 |
3.115 ± 0.031 | 8.966 ± 0.04 |
1.914 ± 0.021 | 4.261 ± 0.031 |
1.739 ± 0.027 | 9.98 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.659 ± 0.015 | 1.871 ± 0.023 |
5.6 ± 0.03 | 2.563 ± 0.021 |
7.577 ± 0.05 | 5.658 ± 0.033 |
6.188 ± 0.038 | 9.211 ± 0.043 |
1.568 ± 0.019 | 1.922 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
