
Hubei permutotetra-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.84
Get precalculated fractions of proteins
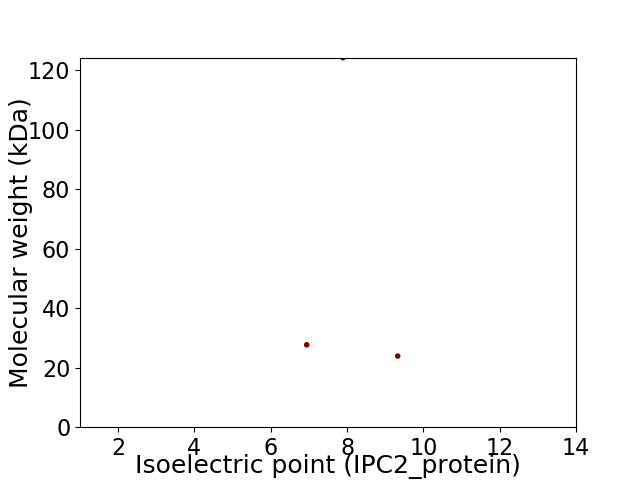
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHQ0|A0A1L3KHQ0_9VIRU RdRp OS=Hubei permutotetra-like virus 4 OX=1923078 PE=4 SV=1
MM1 pKa = 7.51RR2 pKa = 11.84QFSSRR7 pKa = 11.84GRR9 pKa = 11.84GATNNVNARR18 pKa = 11.84NTSGLSHH25 pKa = 6.47TVYY28 pKa = 10.79KK29 pKa = 10.6KK30 pKa = 10.85DD31 pKa = 3.42FTLTTDD37 pKa = 3.1SFTEE41 pKa = 4.46TVSLEE46 pKa = 4.21DD47 pKa = 3.43GGSYY51 pKa = 10.73RR52 pKa = 11.84LIPSVQTEE60 pKa = 4.52TEE62 pKa = 4.34DD63 pKa = 4.83EE64 pKa = 4.76DD65 pKa = 4.64PLDD68 pKa = 4.37PDD70 pKa = 4.14FEE72 pKa = 4.43GLITNIVKK80 pKa = 10.1PPEE83 pKa = 4.12QYY85 pKa = 11.43SNTAHH90 pKa = 6.86LKK92 pKa = 9.95FYY94 pKa = 9.97PLRR97 pKa = 11.84DD98 pKa = 3.27ILKK101 pKa = 9.3FRR103 pKa = 11.84AVDD106 pKa = 3.91FQLNLHH112 pKa = 6.87LSLEE116 pKa = 4.33IVVDD120 pKa = 3.82YY121 pKa = 8.47PTDD124 pKa = 3.6IEE126 pKa = 4.31ALSILWDD133 pKa = 4.59FSQCIYY139 pKa = 10.37NAQVKK144 pKa = 6.95TRR146 pKa = 11.84RR147 pKa = 11.84EE148 pKa = 3.84RR149 pKa = 11.84VEE151 pKa = 3.62GDD153 pKa = 2.78RR154 pKa = 11.84VLFHH158 pKa = 6.76CVAAGLYY165 pKa = 9.94KK166 pKa = 10.03YY167 pKa = 11.01VPVTRR172 pKa = 11.84PFFKK176 pKa = 10.87LSVEE180 pKa = 4.04WTNTGITSSLAYY192 pKa = 9.39TMKK195 pKa = 9.09TQLYY199 pKa = 9.37AFGLFGDD206 pKa = 4.31ARR208 pKa = 11.84QQEE211 pKa = 4.94GFVAPCAMRR220 pKa = 11.84ILKK223 pKa = 9.66KK224 pKa = 10.38RR225 pKa = 11.84PAVAGLTQFSRR236 pKa = 11.84SRR238 pKa = 11.84AMILGYY244 pKa = 10.69
MM1 pKa = 7.51RR2 pKa = 11.84QFSSRR7 pKa = 11.84GRR9 pKa = 11.84GATNNVNARR18 pKa = 11.84NTSGLSHH25 pKa = 6.47TVYY28 pKa = 10.79KK29 pKa = 10.6KK30 pKa = 10.85DD31 pKa = 3.42FTLTTDD37 pKa = 3.1SFTEE41 pKa = 4.46TVSLEE46 pKa = 4.21DD47 pKa = 3.43GGSYY51 pKa = 10.73RR52 pKa = 11.84LIPSVQTEE60 pKa = 4.52TEE62 pKa = 4.34DD63 pKa = 4.83EE64 pKa = 4.76DD65 pKa = 4.64PLDD68 pKa = 4.37PDD70 pKa = 4.14FEE72 pKa = 4.43GLITNIVKK80 pKa = 10.1PPEE83 pKa = 4.12QYY85 pKa = 11.43SNTAHH90 pKa = 6.86LKK92 pKa = 9.95FYY94 pKa = 9.97PLRR97 pKa = 11.84DD98 pKa = 3.27ILKK101 pKa = 9.3FRR103 pKa = 11.84AVDD106 pKa = 3.91FQLNLHH112 pKa = 6.87LSLEE116 pKa = 4.33IVVDD120 pKa = 3.82YY121 pKa = 8.47PTDD124 pKa = 3.6IEE126 pKa = 4.31ALSILWDD133 pKa = 4.59FSQCIYY139 pKa = 10.37NAQVKK144 pKa = 6.95TRR146 pKa = 11.84RR147 pKa = 11.84EE148 pKa = 3.84RR149 pKa = 11.84VEE151 pKa = 3.62GDD153 pKa = 2.78RR154 pKa = 11.84VLFHH158 pKa = 6.76CVAAGLYY165 pKa = 9.94KK166 pKa = 10.03YY167 pKa = 11.01VPVTRR172 pKa = 11.84PFFKK176 pKa = 10.87LSVEE180 pKa = 4.04WTNTGITSSLAYY192 pKa = 9.39TMKK195 pKa = 9.09TQLYY199 pKa = 9.37AFGLFGDD206 pKa = 4.31ARR208 pKa = 11.84QQEE211 pKa = 4.94GFVAPCAMRR220 pKa = 11.84ILKK223 pKa = 9.66KK224 pKa = 10.38RR225 pKa = 11.84PAVAGLTQFSRR236 pKa = 11.84SRR238 pKa = 11.84AMILGYY244 pKa = 10.69
Molecular weight: 27.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI05|A0A1L3KI05_9VIRU Capsid protein OS=Hubei permutotetra-like virus 4 OX=1923078 PE=3 SV=1
MM1 pKa = 7.27TRR3 pKa = 11.84RR4 pKa = 11.84TKK6 pKa = 10.52KK7 pKa = 10.6KK8 pKa = 8.01PTTKK12 pKa = 10.01TMEE15 pKa = 4.37KK16 pKa = 10.4ALVAAMTNLTTKK28 pKa = 10.16RR29 pKa = 11.84RR30 pKa = 11.84APRR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.03GCGAVAEE42 pKa = 4.6GEE44 pKa = 4.19MVLRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 4.02EE51 pKa = 3.99MLVDD55 pKa = 3.55VTLSANTTEE64 pKa = 4.63SNGSVVLALPNFPWLKK80 pKa = 8.76TVAGSFEE87 pKa = 4.1RR88 pKa = 11.84YY89 pKa = 7.3KK90 pKa = 10.22WKK92 pKa = 10.28RR93 pKa = 11.84LNIHH97 pKa = 5.54WRR99 pKa = 11.84AAGGFNKK106 pKa = 10.33GGLVAVGMDD115 pKa = 3.54WSSQMSSAYY124 pKa = 10.4SKK126 pKa = 9.14PTLTAVTEE134 pKa = 4.21RR135 pKa = 11.84QKK137 pKa = 11.25VLSLTPHH144 pKa = 5.96MSLPISANSISKK156 pKa = 9.21MLGLPVRR163 pKa = 11.84MLNSRR168 pKa = 11.84NWYY171 pKa = 9.59DD172 pKa = 3.17ATKK175 pKa = 10.11TDD177 pKa = 4.21DD178 pKa = 3.52EE179 pKa = 5.0GAVGTLRR186 pKa = 11.84YY187 pKa = 8.9AAKK190 pKa = 10.18CDD192 pKa = 3.41SDD194 pKa = 3.75TTAKK198 pKa = 10.5FIGEE202 pKa = 3.71IWVDD206 pKa = 3.62YY207 pKa = 10.72EE208 pKa = 4.51VVLQGTRR215 pKa = 11.84AA216 pKa = 3.18
MM1 pKa = 7.27TRR3 pKa = 11.84RR4 pKa = 11.84TKK6 pKa = 10.52KK7 pKa = 10.6KK8 pKa = 8.01PTTKK12 pKa = 10.01TMEE15 pKa = 4.37KK16 pKa = 10.4ALVAAMTNLTTKK28 pKa = 10.16RR29 pKa = 11.84RR30 pKa = 11.84APRR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.03GCGAVAEE42 pKa = 4.6GEE44 pKa = 4.19MVLRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 4.02EE51 pKa = 3.99MLVDD55 pKa = 3.55VTLSANTTEE64 pKa = 4.63SNGSVVLALPNFPWLKK80 pKa = 8.76TVAGSFEE87 pKa = 4.1RR88 pKa = 11.84YY89 pKa = 7.3KK90 pKa = 10.22WKK92 pKa = 10.28RR93 pKa = 11.84LNIHH97 pKa = 5.54WRR99 pKa = 11.84AAGGFNKK106 pKa = 10.33GGLVAVGMDD115 pKa = 3.54WSSQMSSAYY124 pKa = 10.4SKK126 pKa = 9.14PTLTAVTEE134 pKa = 4.21RR135 pKa = 11.84QKK137 pKa = 11.25VLSLTPHH144 pKa = 5.96MSLPISANSISKK156 pKa = 9.21MLGLPVRR163 pKa = 11.84MLNSRR168 pKa = 11.84NWYY171 pKa = 9.59DD172 pKa = 3.17ATKK175 pKa = 10.11TDD177 pKa = 4.21DD178 pKa = 3.52EE179 pKa = 5.0GAVGTLRR186 pKa = 11.84YY187 pKa = 8.9AAKK190 pKa = 10.18CDD192 pKa = 3.41SDD194 pKa = 3.75TTAKK198 pKa = 10.5FIGEE202 pKa = 3.71IWVDD206 pKa = 3.62YY207 pKa = 10.72EE208 pKa = 4.51VVLQGTRR215 pKa = 11.84AA216 pKa = 3.18
Molecular weight: 23.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1580 |
216 |
1120 |
526.7 |
58.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.608 ± 0.623 | 0.823 ± 0.157 |
5.696 ± 0.485 | 5.696 ± 0.242 |
2.975 ± 0.998 | 6.899 ± 0.401 |
2.089 ± 0.392 | 3.861 ± 0.329 |
5.823 ± 0.478 | 8.671 ± 0.425 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.101 ± 0.454 | 3.354 ± 0.28 |
5.696 ± 0.792 | 3.228 ± 0.396 |
7.025 ± 0.086 | 6.266 ± 0.46 |
7.848 ± 0.75 | 7.595 ± 0.194 |
1.456 ± 0.285 | 3.291 ± 0.366 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
