
Alces alces faeces associated microvirus MP18 4940
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.43
Get precalculated fractions of proteins
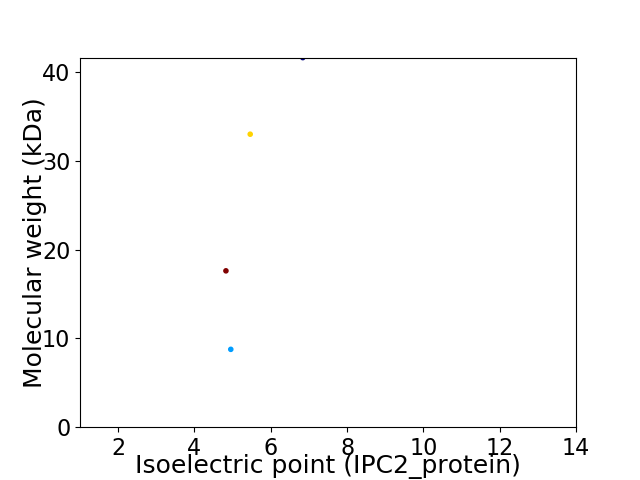
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CL43|A0A2Z5CL43_9VIRU Nonstructural protein OS=Alces alces faeces associated microvirus MP18 4940 OX=2219137 PE=4 SV=1
MM1 pKa = 7.42VKK3 pKa = 10.37FKK5 pKa = 11.07NWFDD9 pKa = 3.22DD10 pKa = 3.88RR11 pKa = 11.84KK12 pKa = 10.3RR13 pKa = 11.84VTSNPGSKK21 pKa = 10.05FVDD24 pKa = 3.27VMQPVVQSDD33 pKa = 3.77GSVEE37 pKa = 3.98LEE39 pKa = 3.93VVGKK43 pKa = 10.17HH44 pKa = 5.38NLYY47 pKa = 11.01DD48 pKa = 4.66EE49 pKa = 4.76IQSHH53 pKa = 5.94ARR55 pKa = 11.84SVDD58 pKa = 2.98INVILQRR65 pKa = 11.84YY66 pKa = 8.76ANGDD70 pKa = 3.58LSALSARR77 pKa = 11.84VPMYY81 pKa = 10.96LDD83 pKa = 3.81LEE85 pKa = 4.49GMPTNYY91 pKa = 9.39MEE93 pKa = 4.44VLNSVIAGEE102 pKa = 4.69KK103 pKa = 10.29FFSDD107 pKa = 3.98LPADD111 pKa = 3.94VKK113 pKa = 11.4SKK115 pKa = 10.84FDD117 pKa = 3.58NNFYY121 pKa = 10.95KK122 pKa = 10.55FVSSIGSDD130 pKa = 3.25EE131 pKa = 4.25FKK133 pKa = 10.97AAFGLKK139 pKa = 10.08SSDD142 pKa = 3.25VVEE145 pKa = 4.3PVVSVEE151 pKa = 3.95KK152 pKa = 10.76EE153 pKa = 4.24GVISEE158 pKa = 4.31
MM1 pKa = 7.42VKK3 pKa = 10.37FKK5 pKa = 11.07NWFDD9 pKa = 3.22DD10 pKa = 3.88RR11 pKa = 11.84KK12 pKa = 10.3RR13 pKa = 11.84VTSNPGSKK21 pKa = 10.05FVDD24 pKa = 3.27VMQPVVQSDD33 pKa = 3.77GSVEE37 pKa = 3.98LEE39 pKa = 3.93VVGKK43 pKa = 10.17HH44 pKa = 5.38NLYY47 pKa = 11.01DD48 pKa = 4.66EE49 pKa = 4.76IQSHH53 pKa = 5.94ARR55 pKa = 11.84SVDD58 pKa = 2.98INVILQRR65 pKa = 11.84YY66 pKa = 8.76ANGDD70 pKa = 3.58LSALSARR77 pKa = 11.84VPMYY81 pKa = 10.96LDD83 pKa = 3.81LEE85 pKa = 4.49GMPTNYY91 pKa = 9.39MEE93 pKa = 4.44VLNSVIAGEE102 pKa = 4.69KK103 pKa = 10.29FFSDD107 pKa = 3.98LPADD111 pKa = 3.94VKK113 pKa = 11.4SKK115 pKa = 10.84FDD117 pKa = 3.58NNFYY121 pKa = 10.95KK122 pKa = 10.55FVSSIGSDD130 pKa = 3.25EE131 pKa = 4.25FKK133 pKa = 10.97AAFGLKK139 pKa = 10.08SSDD142 pKa = 3.25VVEE145 pKa = 4.3PVVSVEE151 pKa = 3.95KK152 pKa = 10.76EE153 pKa = 4.24GVISEE158 pKa = 4.31
Molecular weight: 17.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CIA3|A0A2Z5CIA3_9VIRU Minor capsid protein OS=Alces alces faeces associated microvirus MP18 4940 OX=2219137 PE=4 SV=1
MM1 pKa = 7.66PCYY4 pKa = 10.28HH5 pKa = 7.38PLKK8 pKa = 10.0AFKK11 pKa = 10.9VGITKK16 pKa = 10.38NNKK19 pKa = 9.4DD20 pKa = 3.31KK21 pKa = 11.43LLITSYY27 pKa = 11.04KK28 pKa = 9.86VDD30 pKa = 2.99HH31 pKa = 6.82LEE33 pKa = 4.05YY34 pKa = 10.96NNRR37 pKa = 11.84LNKK40 pKa = 9.57FDD42 pKa = 4.29RR43 pKa = 11.84VYY45 pKa = 11.29SSDD48 pKa = 3.62LVNPYY53 pKa = 9.89ADD55 pKa = 3.76KK56 pKa = 11.15VISCFDD62 pKa = 3.79IPCGHH67 pKa = 7.38CIGCRR72 pKa = 11.84LDD74 pKa = 3.78YY75 pKa = 10.78SRR77 pKa = 11.84NWACRR82 pKa = 11.84MLLEE86 pKa = 4.44VEE88 pKa = 4.38EE89 pKa = 5.12HH90 pKa = 6.0SEE92 pKa = 4.06SWFCTFTYY100 pKa = 10.84DD101 pKa = 3.79NAHH104 pKa = 6.7IPTSVGYY111 pKa = 10.28DD112 pKa = 3.16VDD114 pKa = 4.02SGEE117 pKa = 4.28MFPTVTLRR125 pKa = 11.84KK126 pKa = 9.51RR127 pKa = 11.84DD128 pKa = 3.12WQLFMKK134 pKa = 10.6RR135 pKa = 11.84LRR137 pKa = 11.84KK138 pKa = 9.34WFDD141 pKa = 3.01SVYY144 pKa = 10.33PGEE147 pKa = 4.04KK148 pKa = 9.65LKK150 pKa = 11.21YY151 pKa = 9.2FAAGEE156 pKa = 4.2YY157 pKa = 10.72GDD159 pKa = 3.84TTFRR163 pKa = 11.84PHH165 pKa = 4.73IHH167 pKa = 6.81AIIFGLHH174 pKa = 6.19LPDD177 pKa = 5.12LRR179 pKa = 11.84PSDD182 pKa = 4.01RR183 pKa = 11.84FFSRR187 pKa = 11.84DD188 pKa = 3.22FQYY191 pKa = 11.25YY192 pKa = 9.8EE193 pKa = 4.28SDD195 pKa = 3.07ILSSIWNMGHH205 pKa = 5.47VVVANVTFEE214 pKa = 4.12TCAYY218 pKa = 6.21TARR221 pKa = 11.84YY222 pKa = 8.27IVKK225 pKa = 10.23KK226 pKa = 10.9LNGKK230 pKa = 8.56QADD233 pKa = 4.42LYY235 pKa = 10.44DD236 pKa = 3.36QLGIEE241 pKa = 4.5RR242 pKa = 11.84EE243 pKa = 3.92WALMSRR249 pKa = 11.84KK250 pKa = 9.41PGIGSAYY257 pKa = 9.12ADD259 pKa = 3.71RR260 pKa = 11.84NIEE263 pKa = 3.94HH264 pKa = 7.14LLDD267 pKa = 4.41DD268 pKa = 4.74KK269 pKa = 11.58YY270 pKa = 11.49LIISTEE276 pKa = 4.0DD277 pKa = 3.11GGKK280 pKa = 9.94KK281 pKa = 10.04FPVPNYY287 pKa = 9.46YY288 pKa = 9.89KK289 pKa = 10.52RR290 pKa = 11.84KK291 pKa = 7.75FDD293 pKa = 3.77SWFDD297 pKa = 3.57SQDD300 pKa = 3.33MVEE303 pKa = 4.22SSWYY307 pKa = 10.54KK308 pKa = 10.07FDD310 pKa = 4.16RR311 pKa = 11.84SQAAIRR317 pKa = 11.84VMEE320 pKa = 4.8SIKK323 pKa = 10.16QQQLSNTDD331 pKa = 3.09LSYY334 pKa = 11.11IDD336 pKa = 4.6LLAVNEE342 pKa = 4.23AAQQAKK348 pKa = 9.58MKK350 pKa = 10.36GLEE353 pKa = 4.05RR354 pKa = 11.84KK355 pKa = 8.84VV356 pKa = 3.19
MM1 pKa = 7.66PCYY4 pKa = 10.28HH5 pKa = 7.38PLKK8 pKa = 10.0AFKK11 pKa = 10.9VGITKK16 pKa = 10.38NNKK19 pKa = 9.4DD20 pKa = 3.31KK21 pKa = 11.43LLITSYY27 pKa = 11.04KK28 pKa = 9.86VDD30 pKa = 2.99HH31 pKa = 6.82LEE33 pKa = 4.05YY34 pKa = 10.96NNRR37 pKa = 11.84LNKK40 pKa = 9.57FDD42 pKa = 4.29RR43 pKa = 11.84VYY45 pKa = 11.29SSDD48 pKa = 3.62LVNPYY53 pKa = 9.89ADD55 pKa = 3.76KK56 pKa = 11.15VISCFDD62 pKa = 3.79IPCGHH67 pKa = 7.38CIGCRR72 pKa = 11.84LDD74 pKa = 3.78YY75 pKa = 10.78SRR77 pKa = 11.84NWACRR82 pKa = 11.84MLLEE86 pKa = 4.44VEE88 pKa = 4.38EE89 pKa = 5.12HH90 pKa = 6.0SEE92 pKa = 4.06SWFCTFTYY100 pKa = 10.84DD101 pKa = 3.79NAHH104 pKa = 6.7IPTSVGYY111 pKa = 10.28DD112 pKa = 3.16VDD114 pKa = 4.02SGEE117 pKa = 4.28MFPTVTLRR125 pKa = 11.84KK126 pKa = 9.51RR127 pKa = 11.84DD128 pKa = 3.12WQLFMKK134 pKa = 10.6RR135 pKa = 11.84LRR137 pKa = 11.84KK138 pKa = 9.34WFDD141 pKa = 3.01SVYY144 pKa = 10.33PGEE147 pKa = 4.04KK148 pKa = 9.65LKK150 pKa = 11.21YY151 pKa = 9.2FAAGEE156 pKa = 4.2YY157 pKa = 10.72GDD159 pKa = 3.84TTFRR163 pKa = 11.84PHH165 pKa = 4.73IHH167 pKa = 6.81AIIFGLHH174 pKa = 6.19LPDD177 pKa = 5.12LRR179 pKa = 11.84PSDD182 pKa = 4.01RR183 pKa = 11.84FFSRR187 pKa = 11.84DD188 pKa = 3.22FQYY191 pKa = 11.25YY192 pKa = 9.8EE193 pKa = 4.28SDD195 pKa = 3.07ILSSIWNMGHH205 pKa = 5.47VVVANVTFEE214 pKa = 4.12TCAYY218 pKa = 6.21TARR221 pKa = 11.84YY222 pKa = 8.27IVKK225 pKa = 10.23KK226 pKa = 10.9LNGKK230 pKa = 8.56QADD233 pKa = 4.42LYY235 pKa = 10.44DD236 pKa = 3.36QLGIEE241 pKa = 4.5RR242 pKa = 11.84EE243 pKa = 3.92WALMSRR249 pKa = 11.84KK250 pKa = 9.41PGIGSAYY257 pKa = 9.12ADD259 pKa = 3.71RR260 pKa = 11.84NIEE263 pKa = 3.94HH264 pKa = 7.14LLDD267 pKa = 4.41DD268 pKa = 4.74KK269 pKa = 11.58YY270 pKa = 11.49LIISTEE276 pKa = 4.0DD277 pKa = 3.11GGKK280 pKa = 9.94KK281 pKa = 10.04FPVPNYY287 pKa = 9.46YY288 pKa = 9.89KK289 pKa = 10.52RR290 pKa = 11.84KK291 pKa = 7.75FDD293 pKa = 3.77SWFDD297 pKa = 3.57SQDD300 pKa = 3.33MVEE303 pKa = 4.22SSWYY307 pKa = 10.54KK308 pKa = 10.07FDD310 pKa = 4.16RR311 pKa = 11.84SQAAIRR317 pKa = 11.84VMEE320 pKa = 4.8SIKK323 pKa = 10.16QQQLSNTDD331 pKa = 3.09LSYY334 pKa = 11.11IDD336 pKa = 4.6LLAVNEE342 pKa = 4.23AAQQAKK348 pKa = 9.58MKK350 pKa = 10.36GLEE353 pKa = 4.05RR354 pKa = 11.84KK355 pKa = 8.84VV356 pKa = 3.19
Molecular weight: 41.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
897 |
79 |
356 |
224.3 |
25.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.138 ± 1.988 | 0.892 ± 0.647 |
7.135 ± 0.744 | 5.128 ± 0.616 |
5.24 ± 0.635 | 5.351 ± 0.448 |
1.784 ± 0.552 | 4.348 ± 0.609 |
6.689 ± 0.596 | 6.8 ± 0.665 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.567 ± 0.585 | 5.797 ± 0.894 |
2.676 ± 0.69 | 3.902 ± 0.972 |
4.125 ± 0.851 | 10.479 ± 1.607 |
4.013 ± 0.887 | 7.692 ± 2.343 |
1.449 ± 0.449 | 4.794 ± 0.78 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
