
Torque teno sus virus 1b
Taxonomy: Viruses; Anelloviridae; Iotatorquevirus
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
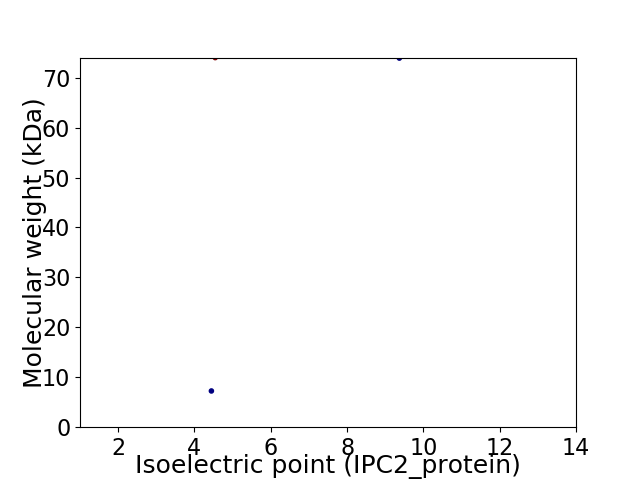
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
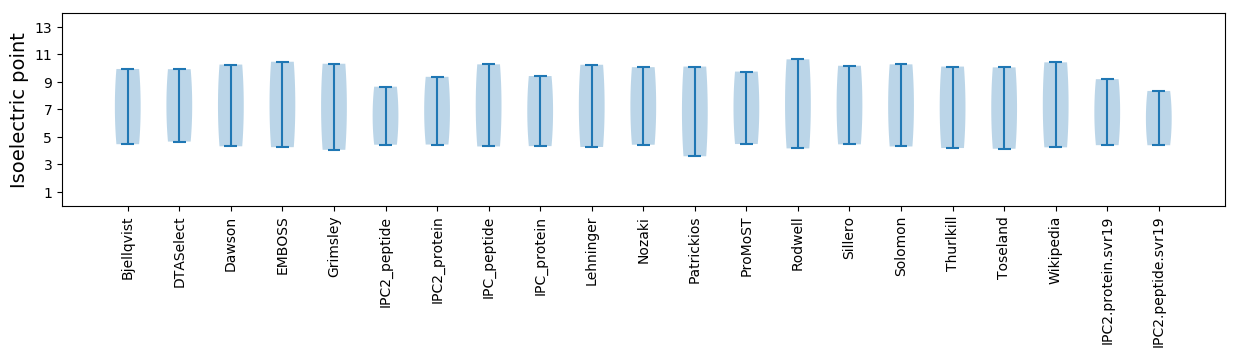
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0Y3X8|E0Y3X8_9VIRU ORF2 OS=Torque teno sus virus 1b OX=687387 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.51EE2 pKa = 4.92EE3 pKa = 3.49RR4 pKa = 11.84WLTVAYY10 pKa = 8.84CAHH13 pKa = 7.1GLFCDD18 pKa = 3.9CKK20 pKa = 10.75DD21 pKa = 3.68PKK23 pKa = 10.89KK24 pKa = 10.17HH25 pKa = 6.64LEE27 pKa = 3.92KK28 pKa = 10.88CLTDD32 pKa = 5.18AIADD36 pKa = 3.82AEE38 pKa = 4.42GDD40 pKa = 3.68RR41 pKa = 11.84QEE43 pKa = 5.44DD44 pKa = 4.1GGTGGGDD51 pKa = 2.92ATFDD55 pKa = 3.25IGIDD59 pKa = 3.51ALLAAAAQRR68 pKa = 3.68
MM1 pKa = 7.51EE2 pKa = 4.92EE3 pKa = 3.49RR4 pKa = 11.84WLTVAYY10 pKa = 8.84CAHH13 pKa = 7.1GLFCDD18 pKa = 3.9CKK20 pKa = 10.75DD21 pKa = 3.68PKK23 pKa = 10.89KK24 pKa = 10.17HH25 pKa = 6.64LEE27 pKa = 3.92KK28 pKa = 10.88CLTDD32 pKa = 5.18AIADD36 pKa = 3.82AEE38 pKa = 4.42GDD40 pKa = 3.68RR41 pKa = 11.84QEE43 pKa = 5.44DD44 pKa = 4.1GGTGGGDD51 pKa = 2.92ATFDD55 pKa = 3.25IGIDD59 pKa = 3.51ALLAAAAQRR68 pKa = 3.68
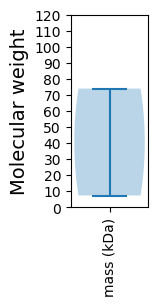
Molecular weight: 7.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0Y3X8|E0Y3X8_9VIRU ORF2 OS=Torque teno sus virus 1b OX=687387 GN=ORF2 PE=4 SV=1
MM1 pKa = 8.01PYY3 pKa = 9.82RR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 9.5RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PTRR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84HH19 pKa = 4.47RR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 7.81FRR27 pKa = 11.84YY28 pKa = 9.49RR29 pKa = 11.84YY30 pKa = 9.15RR31 pKa = 11.84RR32 pKa = 11.84APRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84TKK40 pKa = 8.79VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 10.11APVIQWNPPSRR58 pKa = 11.84RR59 pKa = 11.84TCLIEE64 pKa = 4.59GFWPLSYY71 pKa = 10.62GHH73 pKa = 7.13WFRR76 pKa = 11.84TCLPFRR82 pKa = 11.84RR83 pKa = 11.84KK84 pKa = 10.05NGLIFTGGGCDD95 pKa = 2.95WTQWSLQNLYY105 pKa = 10.4HH106 pKa = 7.36EE107 pKa = 4.67KK108 pKa = 10.89LNWRR112 pKa = 11.84NIWTASNVGMEE123 pKa = 4.38FTRR126 pKa = 11.84FLRR129 pKa = 11.84GKK131 pKa = 9.95FYY133 pKa = 10.73FFRR136 pKa = 11.84HH137 pKa = 5.36PWRR140 pKa = 11.84NYY142 pKa = 8.91IVTWDD147 pKa = 3.48QDD149 pKa = 3.94IPCKK153 pKa = 9.77PLPYY157 pKa = 10.09QNLHH161 pKa = 6.55PLLMLLKK168 pKa = 9.36KK169 pKa = 8.04QHH171 pKa = 6.65KK172 pKa = 9.91LVLSQQNCNPNRR184 pKa = 11.84KK185 pKa = 8.66QKK187 pKa = 10.48PVTLKK192 pKa = 10.25FRR194 pKa = 11.84PPPKK198 pKa = 9.71LTSQWRR204 pKa = 11.84LSRR207 pKa = 11.84EE208 pKa = 3.95LAKK211 pKa = 10.31MPLIRR216 pKa = 11.84LGVSFIDD223 pKa = 4.12LTEE226 pKa = 4.07PWLEE230 pKa = 3.71GWGNAFYY237 pKa = 11.12SVLGYY242 pKa = 9.89EE243 pKa = 4.55AIKK246 pKa = 10.32EE247 pKa = 3.95QGHH250 pKa = 5.32WSNWAQVKK258 pKa = 8.7YY259 pKa = 10.38YY260 pKa = 9.75WIYY263 pKa = 9.93DD264 pKa = 3.69TGVGNAVYY272 pKa = 10.73VVMLKK277 pKa = 10.14QEE279 pKa = 3.97VDD281 pKa = 3.76DD282 pKa = 5.19NPGKK286 pKa = 9.45MASTFKK292 pKa = 8.49TTQGQHH298 pKa = 6.25PNAIDD303 pKa = 4.13HH304 pKa = 6.87IEE306 pKa = 4.66LINEE310 pKa = 4.42GWPYY314 pKa = 10.19WLYY317 pKa = 11.05FFGKK321 pKa = 9.84SEE323 pKa = 3.97QDD325 pKa = 3.06IKK327 pKa = 11.32KK328 pKa = 9.07EE329 pKa = 3.82AHH331 pKa = 5.24SQEE334 pKa = 3.81IARR337 pKa = 11.84EE338 pKa = 3.91YY339 pKa = 9.47ATNPKK344 pKa = 9.84SKK346 pKa = 9.86KK347 pKa = 9.88LKK349 pKa = 9.95IGIVGWASSNFTTPGSSQNTGGNTAAIQGGYY380 pKa = 7.85VAWAGGQGKK389 pKa = 9.92LNLGAGSIGNLYY401 pKa = 8.43QQGWPSNQNWPNTNRR416 pKa = 11.84DD417 pKa = 3.24EE418 pKa = 4.83TNFDD422 pKa = 2.67WGLRR426 pKa = 11.84SLCILRR432 pKa = 11.84DD433 pKa = 3.61NMQLGSQEE441 pKa = 5.12LDD443 pKa = 3.89DD444 pKa = 4.58EE445 pKa = 4.72CTMLSLFGPFVEE457 pKa = 4.54KK458 pKa = 10.79ANPIFATTDD467 pKa = 3.16PKK469 pKa = 11.11YY470 pKa = 10.66FKK472 pKa = 10.64PEE474 pKa = 3.89LKK476 pKa = 10.28DD477 pKa = 3.23YY478 pKa = 10.97NLIMKK483 pKa = 8.98YY484 pKa = 10.38AFKK487 pKa = 10.3FQWGGHH493 pKa = 3.76GTEE496 pKa = 4.41RR497 pKa = 11.84FKK499 pKa = 9.27TTIGDD504 pKa = 3.85PSTIPCPFEE513 pKa = 5.34PGDD516 pKa = 3.9RR517 pKa = 11.84FHH519 pKa = 9.18SGIQDD524 pKa = 3.36PSKK527 pKa = 10.94VQNTVLNPWDD537 pKa = 4.05YY538 pKa = 11.7DD539 pKa = 3.39CDD541 pKa = 4.87GIVRR545 pKa = 11.84KK546 pKa = 8.78DD547 pKa = 3.28TLKK550 pKa = 10.91RR551 pKa = 11.84LLEE554 pKa = 4.48LPTEE558 pKa = 4.31TEE560 pKa = 4.42EE561 pKa = 4.42EE562 pKa = 4.15EE563 pKa = 4.2KK564 pKa = 10.55AYY566 pKa = 9.87PLLGQKK572 pKa = 7.42TEE574 pKa = 4.39KK575 pKa = 10.65EE576 pKa = 4.16PLSDD580 pKa = 3.53SDD582 pKa = 3.95EE583 pKa = 4.33EE584 pKa = 4.55SVISSTSSGSSQEE597 pKa = 4.44EE598 pKa = 3.84EE599 pKa = 4.09TQRR602 pKa = 11.84RR603 pKa = 11.84KK604 pKa = 8.99HH605 pKa = 5.93HH606 pKa = 6.35KK607 pKa = 7.96PSKK610 pKa = 9.9RR611 pKa = 11.84RR612 pKa = 11.84LLKK615 pKa = 10.16HH616 pKa = 5.57LQRR619 pKa = 11.84VVKK622 pKa = 10.41RR623 pKa = 11.84MKK625 pKa = 9.97TLL627 pKa = 3.31
MM1 pKa = 8.01PYY3 pKa = 9.82RR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 9.5RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PTRR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84HH19 pKa = 4.47RR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 7.81FRR27 pKa = 11.84YY28 pKa = 9.49RR29 pKa = 11.84YY30 pKa = 9.15RR31 pKa = 11.84RR32 pKa = 11.84APRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84TKK40 pKa = 8.79VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 10.11APVIQWNPPSRR58 pKa = 11.84RR59 pKa = 11.84TCLIEE64 pKa = 4.59GFWPLSYY71 pKa = 10.62GHH73 pKa = 7.13WFRR76 pKa = 11.84TCLPFRR82 pKa = 11.84RR83 pKa = 11.84KK84 pKa = 10.05NGLIFTGGGCDD95 pKa = 2.95WTQWSLQNLYY105 pKa = 10.4HH106 pKa = 7.36EE107 pKa = 4.67KK108 pKa = 10.89LNWRR112 pKa = 11.84NIWTASNVGMEE123 pKa = 4.38FTRR126 pKa = 11.84FLRR129 pKa = 11.84GKK131 pKa = 9.95FYY133 pKa = 10.73FFRR136 pKa = 11.84HH137 pKa = 5.36PWRR140 pKa = 11.84NYY142 pKa = 8.91IVTWDD147 pKa = 3.48QDD149 pKa = 3.94IPCKK153 pKa = 9.77PLPYY157 pKa = 10.09QNLHH161 pKa = 6.55PLLMLLKK168 pKa = 9.36KK169 pKa = 8.04QHH171 pKa = 6.65KK172 pKa = 9.91LVLSQQNCNPNRR184 pKa = 11.84KK185 pKa = 8.66QKK187 pKa = 10.48PVTLKK192 pKa = 10.25FRR194 pKa = 11.84PPPKK198 pKa = 9.71LTSQWRR204 pKa = 11.84LSRR207 pKa = 11.84EE208 pKa = 3.95LAKK211 pKa = 10.31MPLIRR216 pKa = 11.84LGVSFIDD223 pKa = 4.12LTEE226 pKa = 4.07PWLEE230 pKa = 3.71GWGNAFYY237 pKa = 11.12SVLGYY242 pKa = 9.89EE243 pKa = 4.55AIKK246 pKa = 10.32EE247 pKa = 3.95QGHH250 pKa = 5.32WSNWAQVKK258 pKa = 8.7YY259 pKa = 10.38YY260 pKa = 9.75WIYY263 pKa = 9.93DD264 pKa = 3.69TGVGNAVYY272 pKa = 10.73VVMLKK277 pKa = 10.14QEE279 pKa = 3.97VDD281 pKa = 3.76DD282 pKa = 5.19NPGKK286 pKa = 9.45MASTFKK292 pKa = 8.49TTQGQHH298 pKa = 6.25PNAIDD303 pKa = 4.13HH304 pKa = 6.87IEE306 pKa = 4.66LINEE310 pKa = 4.42GWPYY314 pKa = 10.19WLYY317 pKa = 11.05FFGKK321 pKa = 9.84SEE323 pKa = 3.97QDD325 pKa = 3.06IKK327 pKa = 11.32KK328 pKa = 9.07EE329 pKa = 3.82AHH331 pKa = 5.24SQEE334 pKa = 3.81IARR337 pKa = 11.84EE338 pKa = 3.91YY339 pKa = 9.47ATNPKK344 pKa = 9.84SKK346 pKa = 9.86KK347 pKa = 9.88LKK349 pKa = 9.95IGIVGWASSNFTTPGSSQNTGGNTAAIQGGYY380 pKa = 7.85VAWAGGQGKK389 pKa = 9.92LNLGAGSIGNLYY401 pKa = 8.43QQGWPSNQNWPNTNRR416 pKa = 11.84DD417 pKa = 3.24EE418 pKa = 4.83TNFDD422 pKa = 2.67WGLRR426 pKa = 11.84SLCILRR432 pKa = 11.84DD433 pKa = 3.61NMQLGSQEE441 pKa = 5.12LDD443 pKa = 3.89DD444 pKa = 4.58EE445 pKa = 4.72CTMLSLFGPFVEE457 pKa = 4.54KK458 pKa = 10.79ANPIFATTDD467 pKa = 3.16PKK469 pKa = 11.11YY470 pKa = 10.66FKK472 pKa = 10.64PEE474 pKa = 3.89LKK476 pKa = 10.28DD477 pKa = 3.23YY478 pKa = 10.97NLIMKK483 pKa = 8.98YY484 pKa = 10.38AFKK487 pKa = 10.3FQWGGHH493 pKa = 3.76GTEE496 pKa = 4.41RR497 pKa = 11.84FKK499 pKa = 9.27TTIGDD504 pKa = 3.85PSTIPCPFEE513 pKa = 5.34PGDD516 pKa = 3.9RR517 pKa = 11.84FHH519 pKa = 9.18SGIQDD524 pKa = 3.36PSKK527 pKa = 10.94VQNTVLNPWDD537 pKa = 4.05YY538 pKa = 11.7DD539 pKa = 3.39CDD541 pKa = 4.87GIVRR545 pKa = 11.84KK546 pKa = 8.78DD547 pKa = 3.28TLKK550 pKa = 10.91RR551 pKa = 11.84LLEE554 pKa = 4.48LPTEE558 pKa = 4.31TEE560 pKa = 4.42EE561 pKa = 4.42EE562 pKa = 4.15EE563 pKa = 4.2KK564 pKa = 10.55AYY566 pKa = 9.87PLLGQKK572 pKa = 7.42TEE574 pKa = 4.39KK575 pKa = 10.65EE576 pKa = 4.16PLSDD580 pKa = 3.53SDD582 pKa = 3.95EE583 pKa = 4.33EE584 pKa = 4.55SVISSTSSGSSQEE597 pKa = 4.44EE598 pKa = 3.84EE599 pKa = 4.09TQRR602 pKa = 11.84RR603 pKa = 11.84KK604 pKa = 8.99HH605 pKa = 5.93HH606 pKa = 6.35KK607 pKa = 7.96PSKK610 pKa = 9.9RR611 pKa = 11.84RR612 pKa = 11.84LLKK615 pKa = 10.16HH616 pKa = 5.57LQRR619 pKa = 11.84VVKK622 pKa = 10.41RR623 pKa = 11.84MKK625 pKa = 9.97TLL627 pKa = 3.31
Molecular weight: 73.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
695 |
68 |
627 |
347.5 |
40.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.892 ± 4.87 | 1.871 ± 1.731 |
4.892 ± 3.601 | 5.755 ± 0.689 |
4.173 ± 0.531 | 7.626 ± 1.786 |
2.446 ± 0.214 | 4.317 ± 0.041 |
7.194 ± 0.566 | 8.489 ± 0.144 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.583 ± 0.048 | 4.748 ± 2.049 |
6.043 ± 1.973 | 4.892 ± 0.842 |
8.633 ± 1.822 | 5.18 ± 2.236 |
6.043 ± 0.069 | 3.453 ± 0.856 |
3.885 ± 1.042 | 3.885 ± 1.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
