
Japanese macaque simian foamy virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Simiispumavirus
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
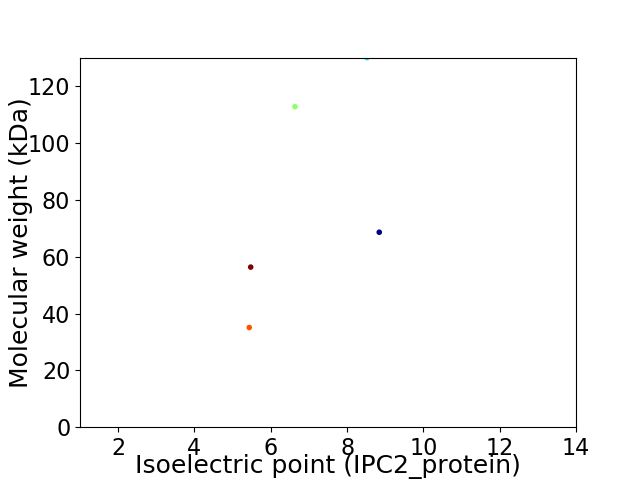
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
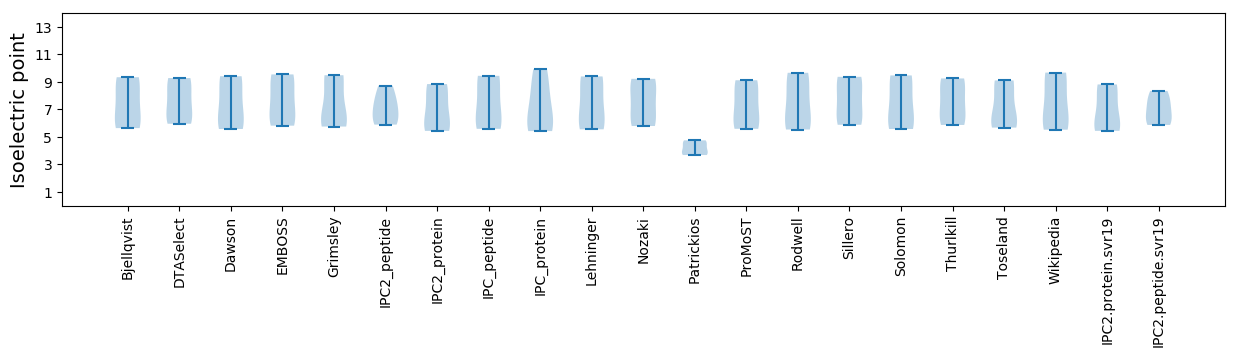
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A077JM51|A0A077JM51_9RETR Bet protein OS=Japanese macaque simian foamy virus OX=2170198 GN=bet PE=4 SV=1
MM1 pKa = 7.61ASWEE5 pKa = 4.14DD6 pKa = 3.63QEE8 pKa = 4.3EE9 pKa = 4.23LRR11 pKa = 11.84EE12 pKa = 4.16LLHH15 pKa = 6.6HH16 pKa = 6.8LPEE19 pKa = 5.62DD20 pKa = 4.48DD21 pKa = 4.43PPADD25 pKa = 4.28LSHH28 pKa = 7.1LLNLDD33 pKa = 3.2EE34 pKa = 5.16MEE36 pKa = 4.49PKK38 pKa = 10.59VLGGQNPGDD47 pKa = 3.54EE48 pKa = 4.71KK49 pKa = 11.08LRR51 pKa = 11.84QQVIKK56 pKa = 9.64PPSIHH61 pKa = 6.98PSTVTWHH68 pKa = 6.54FGYY71 pKa = 10.18KK72 pKa = 10.11DD73 pKa = 3.91KK74 pKa = 11.11EE75 pKa = 4.2DD76 pKa = 3.58QQPEE80 pKa = 3.66IKK82 pKa = 9.91MRR84 pKa = 11.84DD85 pKa = 3.46WVPDD89 pKa = 3.82PSKK92 pKa = 10.33MSKK95 pKa = 7.51STCMRR100 pKa = 11.84LILSSKK106 pKa = 9.82EE107 pKa = 3.97LGSQSVALILAEE119 pKa = 4.08DD120 pKa = 3.97MKK122 pKa = 10.9HH123 pKa = 6.3SEE125 pKa = 5.48SILQLGNGAPRR136 pKa = 11.84KK137 pKa = 9.29GFPLTSSPLLPVVTPWPLSQDD158 pKa = 3.1HH159 pKa = 6.15VAPTLYY165 pKa = 10.66SLLVAYY171 pKa = 8.95YY172 pKa = 10.75KK173 pKa = 10.68SFQNQKK179 pKa = 10.51LEE181 pKa = 4.07PPKK184 pKa = 10.24WLWQCLEE191 pKa = 4.38DD192 pKa = 4.09PSGRR196 pKa = 11.84KK197 pKa = 9.64CMVTQFLVPPLGQVMISCYY216 pKa = 10.45RR217 pKa = 11.84NLTSIIICQAVDD229 pKa = 2.85PWEE232 pKa = 4.37NNNEE236 pKa = 4.03IDD238 pKa = 3.72WKK240 pKa = 9.15KK241 pKa = 11.25DD242 pKa = 2.8PMARR246 pKa = 11.84PRR248 pKa = 11.84IRR250 pKa = 11.84CDD252 pKa = 2.8HH253 pKa = 6.34ALCFKK258 pKa = 10.44VVYY261 pKa = 10.01EE262 pKa = 4.1GTPWRR267 pKa = 11.84PHH269 pKa = 6.0DD270 pKa = 3.74QKK272 pKa = 11.36CWFIRR277 pKa = 11.84LTEE280 pKa = 4.14GHH282 pKa = 5.5KK283 pKa = 10.26HH284 pKa = 5.16GMEE287 pKa = 4.15EE288 pKa = 5.02LSSGDD293 pKa = 3.0WKK295 pKa = 9.72TLQEE299 pKa = 4.25SRR301 pKa = 11.84PYY303 pKa = 9.98PYY305 pKa = 10.74GPVGEE310 pKa = 5.04DD311 pKa = 3.29PNLQYY316 pKa = 11.25AVSVKK321 pKa = 10.03MKK323 pKa = 9.59VTGGPLTSTVLALKK337 pKa = 9.78ALCFHH342 pKa = 7.25RR343 pKa = 11.84VNICNMDD350 pKa = 3.53NPGLGEE356 pKa = 3.8GHH358 pKa = 7.03PPLGYY363 pKa = 10.08SHH365 pKa = 7.23ALKK368 pKa = 10.41AYY370 pKa = 7.68GPQYY374 pKa = 10.2GSCEE378 pKa = 3.91EE379 pKa = 4.53RR380 pKa = 11.84VWQAATKK387 pKa = 10.41CIGPGEE393 pKa = 4.09GDD395 pKa = 3.42YY396 pKa = 10.4WCEE399 pKa = 3.59YY400 pKa = 9.35DD401 pKa = 3.32HH402 pKa = 7.31RR403 pKa = 11.84GYY405 pKa = 11.0FPIIPNKK412 pKa = 10.34LSPTWVRR419 pKa = 11.84HH420 pKa = 4.42AAPYY424 pKa = 9.86GIQRR428 pKa = 11.84LATPYY433 pKa = 10.32DD434 pKa = 3.41LQMFANEE441 pKa = 4.28LLPPGFSINTPSGACYY457 pKa = 9.23ISSRR461 pKa = 11.84RR462 pKa = 11.84LHH464 pKa = 6.19YY465 pKa = 10.94GNEE468 pKa = 4.16GTLQEE473 pKa = 4.2YY474 pKa = 9.94QEE476 pKa = 4.17NCDD479 pKa = 3.82RR480 pKa = 11.84IKK482 pKa = 10.67RR483 pKa = 11.84GYY485 pKa = 10.36EE486 pKa = 4.22DD487 pKa = 3.75ISSSDD492 pKa = 3.61SSDD495 pKa = 3.12EE496 pKa = 4.1DD497 pKa = 3.47
MM1 pKa = 7.61ASWEE5 pKa = 4.14DD6 pKa = 3.63QEE8 pKa = 4.3EE9 pKa = 4.23LRR11 pKa = 11.84EE12 pKa = 4.16LLHH15 pKa = 6.6HH16 pKa = 6.8LPEE19 pKa = 5.62DD20 pKa = 4.48DD21 pKa = 4.43PPADD25 pKa = 4.28LSHH28 pKa = 7.1LLNLDD33 pKa = 3.2EE34 pKa = 5.16MEE36 pKa = 4.49PKK38 pKa = 10.59VLGGQNPGDD47 pKa = 3.54EE48 pKa = 4.71KK49 pKa = 11.08LRR51 pKa = 11.84QQVIKK56 pKa = 9.64PPSIHH61 pKa = 6.98PSTVTWHH68 pKa = 6.54FGYY71 pKa = 10.18KK72 pKa = 10.11DD73 pKa = 3.91KK74 pKa = 11.11EE75 pKa = 4.2DD76 pKa = 3.58QQPEE80 pKa = 3.66IKK82 pKa = 9.91MRR84 pKa = 11.84DD85 pKa = 3.46WVPDD89 pKa = 3.82PSKK92 pKa = 10.33MSKK95 pKa = 7.51STCMRR100 pKa = 11.84LILSSKK106 pKa = 9.82EE107 pKa = 3.97LGSQSVALILAEE119 pKa = 4.08DD120 pKa = 3.97MKK122 pKa = 10.9HH123 pKa = 6.3SEE125 pKa = 5.48SILQLGNGAPRR136 pKa = 11.84KK137 pKa = 9.29GFPLTSSPLLPVVTPWPLSQDD158 pKa = 3.1HH159 pKa = 6.15VAPTLYY165 pKa = 10.66SLLVAYY171 pKa = 8.95YY172 pKa = 10.75KK173 pKa = 10.68SFQNQKK179 pKa = 10.51LEE181 pKa = 4.07PPKK184 pKa = 10.24WLWQCLEE191 pKa = 4.38DD192 pKa = 4.09PSGRR196 pKa = 11.84KK197 pKa = 9.64CMVTQFLVPPLGQVMISCYY216 pKa = 10.45RR217 pKa = 11.84NLTSIIICQAVDD229 pKa = 2.85PWEE232 pKa = 4.37NNNEE236 pKa = 4.03IDD238 pKa = 3.72WKK240 pKa = 9.15KK241 pKa = 11.25DD242 pKa = 2.8PMARR246 pKa = 11.84PRR248 pKa = 11.84IRR250 pKa = 11.84CDD252 pKa = 2.8HH253 pKa = 6.34ALCFKK258 pKa = 10.44VVYY261 pKa = 10.01EE262 pKa = 4.1GTPWRR267 pKa = 11.84PHH269 pKa = 6.0DD270 pKa = 3.74QKK272 pKa = 11.36CWFIRR277 pKa = 11.84LTEE280 pKa = 4.14GHH282 pKa = 5.5KK283 pKa = 10.26HH284 pKa = 5.16GMEE287 pKa = 4.15EE288 pKa = 5.02LSSGDD293 pKa = 3.0WKK295 pKa = 9.72TLQEE299 pKa = 4.25SRR301 pKa = 11.84PYY303 pKa = 9.98PYY305 pKa = 10.74GPVGEE310 pKa = 5.04DD311 pKa = 3.29PNLQYY316 pKa = 11.25AVSVKK321 pKa = 10.03MKK323 pKa = 9.59VTGGPLTSTVLALKK337 pKa = 9.78ALCFHH342 pKa = 7.25RR343 pKa = 11.84VNICNMDD350 pKa = 3.53NPGLGEE356 pKa = 3.8GHH358 pKa = 7.03PPLGYY363 pKa = 10.08SHH365 pKa = 7.23ALKK368 pKa = 10.41AYY370 pKa = 7.68GPQYY374 pKa = 10.2GSCEE378 pKa = 3.91EE379 pKa = 4.53RR380 pKa = 11.84VWQAATKK387 pKa = 10.41CIGPGEE393 pKa = 4.09GDD395 pKa = 3.42YY396 pKa = 10.4WCEE399 pKa = 3.59YY400 pKa = 9.35DD401 pKa = 3.32HH402 pKa = 7.31RR403 pKa = 11.84GYY405 pKa = 11.0FPIIPNKK412 pKa = 10.34LSPTWVRR419 pKa = 11.84HH420 pKa = 4.42AAPYY424 pKa = 9.86GIQRR428 pKa = 11.84LATPYY433 pKa = 10.32DD434 pKa = 3.41LQMFANEE441 pKa = 4.28LLPPGFSINTPSGACYY457 pKa = 9.23ISSRR461 pKa = 11.84RR462 pKa = 11.84LHH464 pKa = 6.19YY465 pKa = 10.94GNEE468 pKa = 4.16GTLQEE473 pKa = 4.2YY474 pKa = 9.94QEE476 pKa = 4.17NCDD479 pKa = 3.82RR480 pKa = 11.84IKK482 pKa = 10.67RR483 pKa = 11.84GYY485 pKa = 10.36EE486 pKa = 4.22DD487 pKa = 3.75ISSSDD492 pKa = 3.61SSDD495 pKa = 3.12EE496 pKa = 4.1DD497 pKa = 3.47
Molecular weight: 56.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077JI63|A0A077JI63_9RETR Env leader protein OS=Japanese macaque simian foamy virus OX=2170198 GN=env PE=4 SV=1
MM1 pKa = 7.41AAIEE5 pKa = 4.57GDD7 pKa = 4.09LDD9 pKa = 3.89VQALTDD15 pKa = 4.0LFNNLGINRR24 pKa = 11.84DD25 pKa = 3.37PRR27 pKa = 11.84HH28 pKa = 6.25RR29 pKa = 11.84EE30 pKa = 3.84VIALRR35 pKa = 11.84MTGGWWGPATRR46 pKa = 11.84YY47 pKa = 10.39NLVTILLQDD56 pKa = 3.94DD57 pKa = 3.72QGQPLQQPRR66 pKa = 11.84WRR68 pKa = 11.84AEE70 pKa = 3.42GRR72 pKa = 11.84AANPAVMFTLEE83 pKa = 5.27APWQDD88 pKa = 3.25LRR90 pKa = 11.84MAFDD94 pKa = 5.12NIDD97 pKa = 3.69LADD100 pKa = 3.81DD101 pKa = 3.75TLRR104 pKa = 11.84FGPLANGNYY113 pKa = 9.57IPGDD117 pKa = 3.71EE118 pKa = 4.48YY119 pKa = 11.41SLEE122 pKa = 4.55FIPPAMQEE130 pKa = 3.74IAQMQRR136 pKa = 11.84DD137 pKa = 3.86EE138 pKa = 4.99LEE140 pKa = 4.03NVLDD144 pKa = 3.67IVGQITMQMSDD155 pKa = 4.6LIGMQDD161 pKa = 3.02AQIRR165 pKa = 11.84GLEE168 pKa = 4.03GQLRR172 pKa = 11.84GLRR175 pKa = 11.84GNLPVAGTPPPPPPSLDD192 pKa = 3.3IQPAAASSPYY202 pKa = 10.13VVPASSAPAAPAAPADD218 pKa = 4.28LGWFAGGPGPGSVDD232 pKa = 2.99PRR234 pKa = 11.84LARR237 pKa = 11.84VAYY240 pKa = 10.38NPFLPGPSDD249 pKa = 3.65GSGVAPVQPSAPPVASPLLPLPPAQPVQPIIQYY282 pKa = 7.09VHH284 pKa = 7.15PPPINPAQQVIPIQHH299 pKa = 5.77IRR301 pKa = 11.84AVTGNAPTNPRR312 pKa = 11.84EE313 pKa = 3.71IPMWIGRR320 pKa = 11.84NASAIEE326 pKa = 4.38GVFPMTTPDD335 pKa = 3.22LRR337 pKa = 11.84CRR339 pKa = 11.84VINALLGGNLGLNLEE354 pKa = 4.46PQHH357 pKa = 6.62CITWASAIATLYY369 pKa = 10.37VRR371 pKa = 11.84THH373 pKa = 5.85GSYY376 pKa = 10.45PIHH379 pKa = 6.36QLAEE383 pKa = 4.1VLRR386 pKa = 11.84GVANSEE392 pKa = 4.06GVAAAYY398 pKa = 9.55QLGMMLTNRR407 pKa = 11.84DD408 pKa = 3.54YY409 pKa = 11.76NLIWGIIRR417 pKa = 11.84PLLPGQAVVTAMQHH431 pKa = 6.74RR432 pKa = 11.84LDD434 pKa = 4.41QEE436 pKa = 4.08VSDD439 pKa = 4.28AARR442 pKa = 11.84IASFINHH449 pKa = 6.58LNGVYY454 pKa = 10.36EE455 pKa = 4.13LLGLNARR462 pKa = 11.84GQSLRR467 pKa = 11.84IPASGGQTTAGTSAGRR483 pKa = 11.84GARR486 pKa = 11.84GRR488 pKa = 11.84RR489 pKa = 11.84SQQGAPGRR497 pKa = 11.84QSSGQSQQQGRR508 pKa = 11.84RR509 pKa = 11.84STQGQSRR516 pKa = 11.84QSDD519 pKa = 3.32NSDD522 pKa = 3.1QNVQRR527 pKa = 11.84QSQGGNGRR535 pKa = 11.84GGYY538 pKa = 7.51NLRR541 pKa = 11.84PRR543 pKa = 11.84TYY545 pKa = 9.41QPQRR549 pKa = 11.84YY550 pKa = 8.66GGGRR554 pKa = 11.84GRR556 pKa = 11.84RR557 pKa = 11.84WNDD560 pKa = 2.85QPARR564 pKa = 11.84SDD566 pKa = 3.33NQQRR570 pKa = 11.84SQSQQPQSEE579 pKa = 4.15ARR581 pKa = 11.84GEE583 pKa = 3.9QSRR586 pKa = 11.84TSGAGRR592 pKa = 11.84GQGGRR597 pKa = 11.84GNQNRR602 pKa = 11.84NQRR605 pKa = 11.84LAGGNADD612 pKa = 3.5RR613 pKa = 11.84TVNTVTTASASTSASGQDD631 pKa = 3.74GSSPAPPASGSGNN644 pKa = 3.32
MM1 pKa = 7.41AAIEE5 pKa = 4.57GDD7 pKa = 4.09LDD9 pKa = 3.89VQALTDD15 pKa = 4.0LFNNLGINRR24 pKa = 11.84DD25 pKa = 3.37PRR27 pKa = 11.84HH28 pKa = 6.25RR29 pKa = 11.84EE30 pKa = 3.84VIALRR35 pKa = 11.84MTGGWWGPATRR46 pKa = 11.84YY47 pKa = 10.39NLVTILLQDD56 pKa = 3.94DD57 pKa = 3.72QGQPLQQPRR66 pKa = 11.84WRR68 pKa = 11.84AEE70 pKa = 3.42GRR72 pKa = 11.84AANPAVMFTLEE83 pKa = 5.27APWQDD88 pKa = 3.25LRR90 pKa = 11.84MAFDD94 pKa = 5.12NIDD97 pKa = 3.69LADD100 pKa = 3.81DD101 pKa = 3.75TLRR104 pKa = 11.84FGPLANGNYY113 pKa = 9.57IPGDD117 pKa = 3.71EE118 pKa = 4.48YY119 pKa = 11.41SLEE122 pKa = 4.55FIPPAMQEE130 pKa = 3.74IAQMQRR136 pKa = 11.84DD137 pKa = 3.86EE138 pKa = 4.99LEE140 pKa = 4.03NVLDD144 pKa = 3.67IVGQITMQMSDD155 pKa = 4.6LIGMQDD161 pKa = 3.02AQIRR165 pKa = 11.84GLEE168 pKa = 4.03GQLRR172 pKa = 11.84GLRR175 pKa = 11.84GNLPVAGTPPPPPPSLDD192 pKa = 3.3IQPAAASSPYY202 pKa = 10.13VVPASSAPAAPAAPADD218 pKa = 4.28LGWFAGGPGPGSVDD232 pKa = 2.99PRR234 pKa = 11.84LARR237 pKa = 11.84VAYY240 pKa = 10.38NPFLPGPSDD249 pKa = 3.65GSGVAPVQPSAPPVASPLLPLPPAQPVQPIIQYY282 pKa = 7.09VHH284 pKa = 7.15PPPINPAQQVIPIQHH299 pKa = 5.77IRR301 pKa = 11.84AVTGNAPTNPRR312 pKa = 11.84EE313 pKa = 3.71IPMWIGRR320 pKa = 11.84NASAIEE326 pKa = 4.38GVFPMTTPDD335 pKa = 3.22LRR337 pKa = 11.84CRR339 pKa = 11.84VINALLGGNLGLNLEE354 pKa = 4.46PQHH357 pKa = 6.62CITWASAIATLYY369 pKa = 10.37VRR371 pKa = 11.84THH373 pKa = 5.85GSYY376 pKa = 10.45PIHH379 pKa = 6.36QLAEE383 pKa = 4.1VLRR386 pKa = 11.84GVANSEE392 pKa = 4.06GVAAAYY398 pKa = 9.55QLGMMLTNRR407 pKa = 11.84DD408 pKa = 3.54YY409 pKa = 11.76NLIWGIIRR417 pKa = 11.84PLLPGQAVVTAMQHH431 pKa = 6.74RR432 pKa = 11.84LDD434 pKa = 4.41QEE436 pKa = 4.08VSDD439 pKa = 4.28AARR442 pKa = 11.84IASFINHH449 pKa = 6.58LNGVYY454 pKa = 10.36EE455 pKa = 4.13LLGLNARR462 pKa = 11.84GQSLRR467 pKa = 11.84IPASGGQTTAGTSAGRR483 pKa = 11.84GARR486 pKa = 11.84GRR488 pKa = 11.84RR489 pKa = 11.84SQQGAPGRR497 pKa = 11.84QSSGQSQQQGRR508 pKa = 11.84RR509 pKa = 11.84STQGQSRR516 pKa = 11.84QSDD519 pKa = 3.32NSDD522 pKa = 3.1QNVQRR527 pKa = 11.84QSQGGNGRR535 pKa = 11.84GGYY538 pKa = 7.51NLRR541 pKa = 11.84PRR543 pKa = 11.84TYY545 pKa = 9.41QPQRR549 pKa = 11.84YY550 pKa = 8.66GGGRR554 pKa = 11.84GRR556 pKa = 11.84RR557 pKa = 11.84WNDD560 pKa = 2.85QPARR564 pKa = 11.84SDD566 pKa = 3.33NQQRR570 pKa = 11.84SQSQQPQSEE579 pKa = 4.15ARR581 pKa = 11.84GEE583 pKa = 3.9QSRR586 pKa = 11.84TSGAGRR592 pKa = 11.84GQGGRR597 pKa = 11.84GNQNRR602 pKa = 11.84NQRR605 pKa = 11.84LAGGNADD612 pKa = 3.5RR613 pKa = 11.84TVNTVTTASASTSASGQDD631 pKa = 3.74GSSPAPPASGSGNN644 pKa = 3.32
Molecular weight: 68.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3579 |
308 |
1149 |
715.8 |
80.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.259 ± 0.999 | 1.565 ± 0.523 |
4.75 ± 0.348 | 5.644 ± 0.708 |
2.459 ± 0.284 | 6.007 ± 1.299 |
2.626 ± 0.317 | 5.979 ± 0.629 |
5.56 ± 1.3 | 9.835 ± 0.476 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.151 ± 0.261 | 4.554 ± 0.536 |
7.516 ± 0.992 | 6.147 ± 0.666 |
4.973 ± 0.781 | 7.097 ± 0.189 |
5.337 ± 0.512 | 5.7 ± 0.406 |
1.928 ± 0.178 | 3.912 ± 0.514 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
