
Cacao virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Cacao phlebovirus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
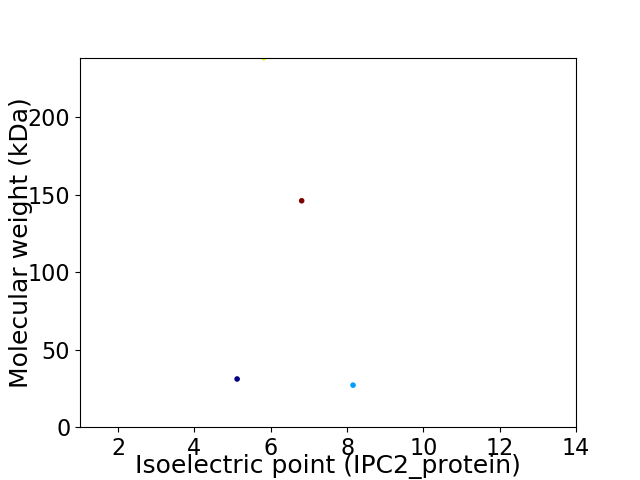
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8D7U2|A0A4P8D7U2_9VIRU Glycoprotein OS=Cacao virus OX=629730 PE=4 SV=1
MM1 pKa = 7.58NYY3 pKa = 10.75YY4 pKa = 9.46MLDD7 pKa = 3.6MPLVSYY13 pKa = 8.97STLNIRR19 pKa = 11.84RR20 pKa = 11.84IQVDD24 pKa = 3.44YY25 pKa = 11.66VPFNKK30 pKa = 9.62MSVCPVSKK38 pKa = 10.91YY39 pKa = 10.35EE40 pKa = 4.04DD41 pKa = 3.31MEE43 pKa = 4.71FEE45 pKa = 4.42VTSYY49 pKa = 10.09SQKK52 pKa = 10.23PGSKK56 pKa = 8.26NTLYY60 pKa = 10.89DD61 pKa = 4.49FYY63 pKa = 11.32SVGEE67 pKa = 4.33IPVSWGVNHH76 pKa = 6.91PGVRR80 pKa = 11.84LEE82 pKa = 4.18SSTKK86 pKa = 9.47FLSLIHH92 pKa = 6.45EE93 pKa = 4.66ISRR96 pKa = 11.84LEE98 pKa = 3.86INDD101 pKa = 3.53VLKK104 pKa = 11.12HH105 pKa = 5.77NEE107 pKa = 4.05PNLKK111 pKa = 10.0RR112 pKa = 11.84ALCWPFSYY120 pKa = 8.14PTLQFFSLIRR130 pKa = 11.84LKK132 pKa = 11.11DD133 pKa = 3.55EE134 pKa = 4.83LGPWSYY140 pKa = 11.27KK141 pKa = 10.31GIVMTQIMRR150 pKa = 11.84ASKK153 pKa = 8.87CTQIDD158 pKa = 3.45RR159 pKa = 11.84CVVNFHH165 pKa = 6.76KK166 pKa = 10.72RR167 pKa = 11.84IQRR170 pKa = 11.84EE171 pKa = 3.98ALEE174 pKa = 4.72LGDD177 pKa = 6.49DD178 pKa = 3.76IGCYY182 pKa = 9.45PGRR185 pKa = 11.84NLVLEE190 pKa = 4.56ACSVICLRR198 pKa = 11.84LLQSYY203 pKa = 9.8RR204 pKa = 11.84YY205 pKa = 9.32DD206 pKa = 3.61LEE208 pKa = 5.16YY209 pKa = 10.86DD210 pKa = 3.67APHH213 pKa = 6.45SKK215 pKa = 10.5LMKK218 pKa = 10.19LVQDD222 pKa = 4.0FDD224 pKa = 4.08EE225 pKa = 5.04SFEE228 pKa = 4.29LVSDD232 pKa = 3.78ADD234 pKa = 3.49LRR236 pKa = 11.84MEE238 pKa = 4.23VARR241 pKa = 11.84DD242 pKa = 3.44EE243 pKa = 4.97FDD245 pKa = 3.8TIMKK249 pKa = 10.02RR250 pKa = 11.84RR251 pKa = 11.84EE252 pKa = 3.92AEE254 pKa = 3.89EE255 pKa = 4.36TFWGSDD261 pKa = 3.28MTDD264 pKa = 2.69SDD266 pKa = 3.92
MM1 pKa = 7.58NYY3 pKa = 10.75YY4 pKa = 9.46MLDD7 pKa = 3.6MPLVSYY13 pKa = 8.97STLNIRR19 pKa = 11.84RR20 pKa = 11.84IQVDD24 pKa = 3.44YY25 pKa = 11.66VPFNKK30 pKa = 9.62MSVCPVSKK38 pKa = 10.91YY39 pKa = 10.35EE40 pKa = 4.04DD41 pKa = 3.31MEE43 pKa = 4.71FEE45 pKa = 4.42VTSYY49 pKa = 10.09SQKK52 pKa = 10.23PGSKK56 pKa = 8.26NTLYY60 pKa = 10.89DD61 pKa = 4.49FYY63 pKa = 11.32SVGEE67 pKa = 4.33IPVSWGVNHH76 pKa = 6.91PGVRR80 pKa = 11.84LEE82 pKa = 4.18SSTKK86 pKa = 9.47FLSLIHH92 pKa = 6.45EE93 pKa = 4.66ISRR96 pKa = 11.84LEE98 pKa = 3.86INDD101 pKa = 3.53VLKK104 pKa = 11.12HH105 pKa = 5.77NEE107 pKa = 4.05PNLKK111 pKa = 10.0RR112 pKa = 11.84ALCWPFSYY120 pKa = 8.14PTLQFFSLIRR130 pKa = 11.84LKK132 pKa = 11.11DD133 pKa = 3.55EE134 pKa = 4.83LGPWSYY140 pKa = 11.27KK141 pKa = 10.31GIVMTQIMRR150 pKa = 11.84ASKK153 pKa = 8.87CTQIDD158 pKa = 3.45RR159 pKa = 11.84CVVNFHH165 pKa = 6.76KK166 pKa = 10.72RR167 pKa = 11.84IQRR170 pKa = 11.84EE171 pKa = 3.98ALEE174 pKa = 4.72LGDD177 pKa = 6.49DD178 pKa = 3.76IGCYY182 pKa = 9.45PGRR185 pKa = 11.84NLVLEE190 pKa = 4.56ACSVICLRR198 pKa = 11.84LLQSYY203 pKa = 9.8RR204 pKa = 11.84YY205 pKa = 9.32DD206 pKa = 3.61LEE208 pKa = 5.16YY209 pKa = 10.86DD210 pKa = 3.67APHH213 pKa = 6.45SKK215 pKa = 10.5LMKK218 pKa = 10.19LVQDD222 pKa = 4.0FDD224 pKa = 4.08EE225 pKa = 5.04SFEE228 pKa = 4.29LVSDD232 pKa = 3.78ADD234 pKa = 3.49LRR236 pKa = 11.84MEE238 pKa = 4.23VARR241 pKa = 11.84DD242 pKa = 3.44EE243 pKa = 4.97FDD245 pKa = 3.8TIMKK249 pKa = 10.02RR250 pKa = 11.84RR251 pKa = 11.84EE252 pKa = 3.92AEE254 pKa = 3.89EE255 pKa = 4.36TFWGSDD261 pKa = 3.28MTDD264 pKa = 2.69SDD266 pKa = 3.92
Molecular weight: 31.09 kDa
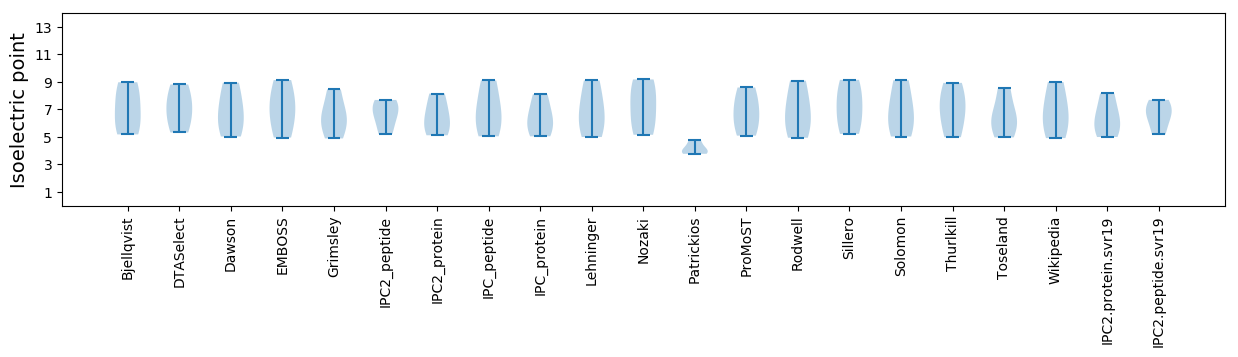
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8D7S5|A0A4P8D7S5_9VIRU NSs OS=Cacao virus OX=629730 PE=4 SV=1
MM1 pKa = 7.97SDD3 pKa = 3.59YY4 pKa = 10.9QQLAVEE10 pKa = 5.11FASEE14 pKa = 4.43VPSEE18 pKa = 4.03AVIATWVEE26 pKa = 3.72DD27 pKa = 3.7FAYY30 pKa = 10.38QGFDD34 pKa = 2.79AGEE37 pKa = 4.1VIKK40 pKa = 11.03ALVEE44 pKa = 3.97KK45 pKa = 10.61GKK47 pKa = 10.33NDD49 pKa = 3.22WKK51 pKa = 10.65QDD53 pKa = 3.21ARR55 pKa = 11.84KK56 pKa = 8.99MIILSLTRR64 pKa = 11.84GNKK67 pKa = 7.45PSKK70 pKa = 7.64MTEE73 pKa = 3.73RR74 pKa = 11.84MSDD77 pKa = 3.11AGKK80 pKa = 8.58STVNALVKK88 pKa = 10.26RR89 pKa = 11.84YY90 pKa = 8.35NLKK93 pKa = 10.47SGNPGRR99 pKa = 11.84KK100 pKa = 9.28DD101 pKa = 3.21LTLSRR106 pKa = 11.84IATALAGWTCQAAPIVQDD124 pKa = 3.62YY125 pKa = 11.15LPVTGKK131 pKa = 11.16SMDD134 pKa = 4.11LLSNNYY140 pKa = 8.09PRR142 pKa = 11.84QMMHH146 pKa = 6.72PCFSGLIDD154 pKa = 3.5PTLPKK159 pKa = 9.94NTVDD163 pKa = 3.5NLSFAHH169 pKa = 6.49CLYY172 pKa = 7.95MTQFSKK178 pKa = 9.97TINPSLRR185 pKa = 11.84GLSKK189 pKa = 10.98SEE191 pKa = 4.04VVSSFSQPMNAAINSSFLSSDD212 pKa = 3.01QRR214 pKa = 11.84RR215 pKa = 11.84SFLDD219 pKa = 3.23TLGLLNKK226 pKa = 10.06NLEE229 pKa = 4.44CTSQVIDD236 pKa = 3.39AAKK239 pKa = 10.43AFKK242 pKa = 10.86SLTNN246 pKa = 3.71
MM1 pKa = 7.97SDD3 pKa = 3.59YY4 pKa = 10.9QQLAVEE10 pKa = 5.11FASEE14 pKa = 4.43VPSEE18 pKa = 4.03AVIATWVEE26 pKa = 3.72DD27 pKa = 3.7FAYY30 pKa = 10.38QGFDD34 pKa = 2.79AGEE37 pKa = 4.1VIKK40 pKa = 11.03ALVEE44 pKa = 3.97KK45 pKa = 10.61GKK47 pKa = 10.33NDD49 pKa = 3.22WKK51 pKa = 10.65QDD53 pKa = 3.21ARR55 pKa = 11.84KK56 pKa = 8.99MIILSLTRR64 pKa = 11.84GNKK67 pKa = 7.45PSKK70 pKa = 7.64MTEE73 pKa = 3.73RR74 pKa = 11.84MSDD77 pKa = 3.11AGKK80 pKa = 8.58STVNALVKK88 pKa = 10.26RR89 pKa = 11.84YY90 pKa = 8.35NLKK93 pKa = 10.47SGNPGRR99 pKa = 11.84KK100 pKa = 9.28DD101 pKa = 3.21LTLSRR106 pKa = 11.84IATALAGWTCQAAPIVQDD124 pKa = 3.62YY125 pKa = 11.15LPVTGKK131 pKa = 11.16SMDD134 pKa = 4.11LLSNNYY140 pKa = 8.09PRR142 pKa = 11.84QMMHH146 pKa = 6.72PCFSGLIDD154 pKa = 3.5PTLPKK159 pKa = 9.94NTVDD163 pKa = 3.5NLSFAHH169 pKa = 6.49CLYY172 pKa = 7.95MTQFSKK178 pKa = 9.97TINPSLRR185 pKa = 11.84GLSKK189 pKa = 10.98SEE191 pKa = 4.04VVSSFSQPMNAAINSSFLSSDD212 pKa = 3.01QRR214 pKa = 11.84RR215 pKa = 11.84SFLDD219 pKa = 3.23TLGLLNKK226 pKa = 10.06NLEE229 pKa = 4.44CTSQVIDD236 pKa = 3.39AAKK239 pKa = 10.43AFKK242 pKa = 10.86SLTNN246 pKa = 3.71
Molecular weight: 27.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3911 |
246 |
2090 |
977.8 |
110.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.5 ± 0.556 | 3.094 ± 0.662 |
5.702 ± 0.402 | 6.52 ± 0.656 |
4.296 ± 0.287 | 5.574 ± 0.451 |
2.199 ± 0.246 | 7.006 ± 0.579 |
6.622 ± 0.241 | 9.0 ± 0.283 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.324 ± 0.191 | 4.96 ± 0.587 |
3.554 ± 0.259 | 3.298 ± 0.15 |
4.986 ± 0.27 | 9.946 ± 0.166 |
5.318 ± 0.206 | 5.651 ± 0.226 |
1.176 ± 0.127 | 3.273 ± 0.272 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
