
Acanthocheilonema viteae (Filarial nematode worm) (Dipetalonema viteae)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Spirurina; Spiruromorpha; Filarioidea; Onchocercidae; Acanthocheilonema
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
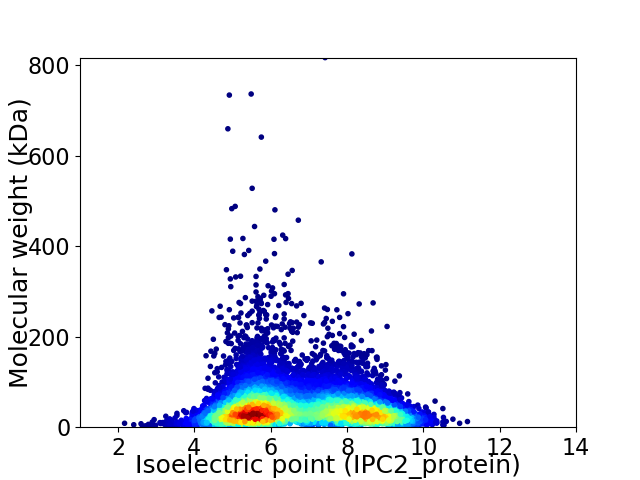
Virtual 2D-PAGE plot for 10007 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A498S4L9|A0A498S4L9_ACAVI Methylenetetrahydrofolate reductase OS=Acanthocheilonema viteae OX=6277 GN=NAV_LOCUS435 PE=3 SV=1
MM1 pKa = 7.76LGLQSSNNRR10 pKa = 11.84EE11 pKa = 3.84EE12 pKa = 5.06PIVKK16 pKa = 9.86SSKK19 pKa = 10.42SALTLQYY26 pKa = 11.16AEE28 pKa = 4.12QCPTCRR34 pKa = 11.84THH36 pKa = 7.14TSPSNIVKK44 pKa = 10.33RR45 pKa = 11.84LYY47 pKa = 10.09FANCDD52 pKa = 3.63DD53 pKa = 3.87KK54 pKa = 11.42TVNDD58 pKa = 3.95VGTSSIAAVTDD69 pKa = 3.87HH70 pKa = 6.59NKK72 pKa = 10.13SASITNEE79 pKa = 3.52NARR82 pKa = 11.84SSQAPEE88 pKa = 4.36NINEE92 pKa = 3.97FDD94 pKa = 4.76GNYY97 pKa = 9.79HH98 pKa = 7.66DD99 pKa = 5.59NGKK102 pKa = 10.16NYY104 pKa = 10.33NNNSGYY110 pKa = 10.92DD111 pKa = 3.25SDD113 pKa = 5.8YY114 pKa = 11.17EE115 pKa = 4.33DD116 pKa = 3.66EE117 pKa = 4.54ANRR120 pKa = 11.84AIDD123 pKa = 4.33GDD125 pKa = 4.12FTDD128 pKa = 5.38SSDD131 pKa = 4.94LNMTPSEE138 pKa = 4.13DD139 pKa = 3.79SEE141 pKa = 4.38FATEE145 pKa = 4.84SEE147 pKa = 4.53DD148 pKa = 3.68KK149 pKa = 11.31NEE151 pKa = 3.78EE152 pKa = 3.94LEE154 pKa = 4.17YY155 pKa = 11.1QEE157 pKa = 5.15TISDD161 pKa = 3.79SDD163 pKa = 3.95VVSVLNAPLSITSSVSLGSFAEE185 pKa = 4.36SFGEE189 pKa = 4.11IDD191 pKa = 3.67DD192 pKa = 4.21TLSVSSLEE200 pKa = 3.92YY201 pKa = 11.11DD202 pKa = 3.09NMEE205 pKa = 4.02ISEE208 pKa = 4.49SLDD211 pKa = 3.25DD212 pKa = 4.98HH213 pKa = 8.36DD214 pKa = 5.85EE215 pKa = 4.12ISNDD219 pKa = 2.83GDD221 pKa = 3.92NIFTSGLLIEE231 pKa = 5.42HH232 pKa = 7.57DD233 pKa = 4.62SNDD236 pKa = 3.75DD237 pKa = 3.84DD238 pKa = 5.62SFSSDD243 pKa = 4.41DD244 pKa = 3.87SDD246 pKa = 4.64NGNSDD251 pKa = 3.67SDD253 pKa = 4.23LYY255 pKa = 11.42NYY257 pKa = 11.05DD258 pKa = 2.98HH259 pKa = 7.17FAPHH263 pKa = 6.58LRR265 pKa = 11.84MFNQFILPYY274 pKa = 9.85PMYY277 pKa = 10.62YY278 pKa = 10.24DD279 pKa = 3.5VRR281 pKa = 11.84FQFLFNN287 pKa = 4.07
MM1 pKa = 7.76LGLQSSNNRR10 pKa = 11.84EE11 pKa = 3.84EE12 pKa = 5.06PIVKK16 pKa = 9.86SSKK19 pKa = 10.42SALTLQYY26 pKa = 11.16AEE28 pKa = 4.12QCPTCRR34 pKa = 11.84THH36 pKa = 7.14TSPSNIVKK44 pKa = 10.33RR45 pKa = 11.84LYY47 pKa = 10.09FANCDD52 pKa = 3.63DD53 pKa = 3.87KK54 pKa = 11.42TVNDD58 pKa = 3.95VGTSSIAAVTDD69 pKa = 3.87HH70 pKa = 6.59NKK72 pKa = 10.13SASITNEE79 pKa = 3.52NARR82 pKa = 11.84SSQAPEE88 pKa = 4.36NINEE92 pKa = 3.97FDD94 pKa = 4.76GNYY97 pKa = 9.79HH98 pKa = 7.66DD99 pKa = 5.59NGKK102 pKa = 10.16NYY104 pKa = 10.33NNNSGYY110 pKa = 10.92DD111 pKa = 3.25SDD113 pKa = 5.8YY114 pKa = 11.17EE115 pKa = 4.33DD116 pKa = 3.66EE117 pKa = 4.54ANRR120 pKa = 11.84AIDD123 pKa = 4.33GDD125 pKa = 4.12FTDD128 pKa = 5.38SSDD131 pKa = 4.94LNMTPSEE138 pKa = 4.13DD139 pKa = 3.79SEE141 pKa = 4.38FATEE145 pKa = 4.84SEE147 pKa = 4.53DD148 pKa = 3.68KK149 pKa = 11.31NEE151 pKa = 3.78EE152 pKa = 3.94LEE154 pKa = 4.17YY155 pKa = 11.1QEE157 pKa = 5.15TISDD161 pKa = 3.79SDD163 pKa = 3.95VVSVLNAPLSITSSVSLGSFAEE185 pKa = 4.36SFGEE189 pKa = 4.11IDD191 pKa = 3.67DD192 pKa = 4.21TLSVSSLEE200 pKa = 3.92YY201 pKa = 11.11DD202 pKa = 3.09NMEE205 pKa = 4.02ISEE208 pKa = 4.49SLDD211 pKa = 3.25DD212 pKa = 4.98HH213 pKa = 8.36DD214 pKa = 5.85EE215 pKa = 4.12ISNDD219 pKa = 2.83GDD221 pKa = 3.92NIFTSGLLIEE231 pKa = 5.42HH232 pKa = 7.57DD233 pKa = 4.62SNDD236 pKa = 3.75DD237 pKa = 3.84DD238 pKa = 5.62SFSSDD243 pKa = 4.41DD244 pKa = 3.87SDD246 pKa = 4.64NGNSDD251 pKa = 3.67SDD253 pKa = 4.23LYY255 pKa = 11.42NYY257 pKa = 11.05DD258 pKa = 2.98HH259 pKa = 7.17FAPHH263 pKa = 6.58LRR265 pKa = 11.84MFNQFILPYY274 pKa = 9.85PMYY277 pKa = 10.62YY278 pKa = 10.24DD279 pKa = 3.5VRR281 pKa = 11.84FQFLFNN287 pKa = 4.07
Molecular weight: 32.02 kDa
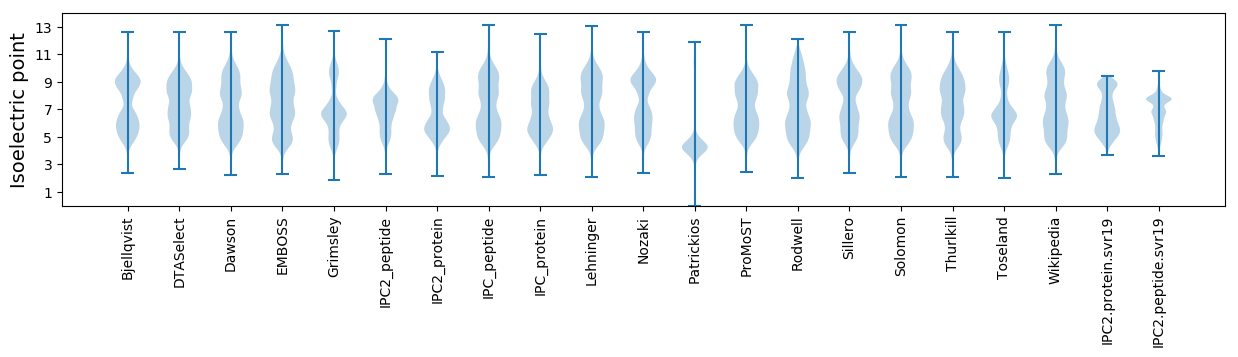
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A498SH36|A0A498SH36_ACAVI Uncharacterized protein OS=Acanthocheilonema viteae OX=6277 GN=NAV_LOCUS4068 PE=4 SV=1
MM1 pKa = 8.08PYY3 pKa = 9.69RR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84SVSRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84SRR16 pKa = 11.84SRR18 pKa = 11.84PSRR21 pKa = 11.84DD22 pKa = 3.43VISLSLVPSRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84SLSQSLLGYY47 pKa = 10.34DD48 pKa = 4.32LLRR51 pKa = 11.84SRR53 pKa = 11.84SPRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84SYY60 pKa = 9.22IRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84SSSRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84SRR74 pKa = 11.84SRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84ILPRR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84LSRR87 pKa = 11.84PRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84SRR93 pKa = 11.84SGYY96 pKa = 9.73RR97 pKa = 11.84YY98 pKa = 9.73RR99 pKa = 5.31
MM1 pKa = 8.08PYY3 pKa = 9.69RR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84SVSRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84SRR16 pKa = 11.84SRR18 pKa = 11.84PSRR21 pKa = 11.84DD22 pKa = 3.43VISLSLVPSRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84SLSQSLLGYY47 pKa = 10.34DD48 pKa = 4.32LLRR51 pKa = 11.84SRR53 pKa = 11.84SPRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84SYY60 pKa = 9.22IRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84SSSRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84SRR74 pKa = 11.84SRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84ILPRR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84LSRR87 pKa = 11.84PRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84SRR93 pKa = 11.84SGYY96 pKa = 9.73RR97 pKa = 11.84YY98 pKa = 9.73RR99 pKa = 5.31
Molecular weight: 12.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4369685 |
19 |
7164 |
436.7 |
49.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.255 ± 0.021 | 2.171 ± 0.022 |
5.531 ± 0.019 | 6.722 ± 0.025 |
4.081 ± 0.018 | 5.154 ± 0.028 |
2.383 ± 0.011 | 6.523 ± 0.026 |
6.172 ± 0.025 | 9.212 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.487 ± 0.009 | 5.127 ± 0.018 |
4.343 ± 0.024 | 4.167 ± 0.024 |
5.764 ± 0.023 | 8.166 ± 0.029 |
5.57 ± 0.017 | 5.899 ± 0.019 |
1.098 ± 0.008 | 3.151 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
