
Desulfuromonas sp. DDH964
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfuromonadales; Desulfuromonadaceae; Desulfuromonas; unclassified Desulfuromonas
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
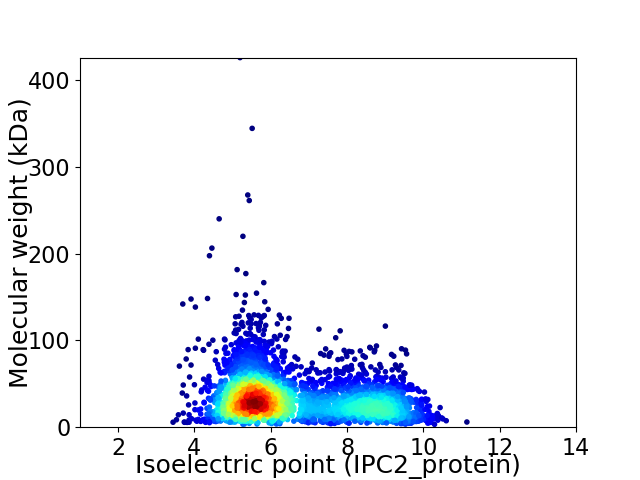
Virtual 2D-PAGE plot for 3483 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A143B473|A0A143B473_9DELT UbiD family decarboxylase OS=Desulfuromonas sp. DDH964 OX=1823759 GN=DBW_0493 PE=4 SV=1
MM1 pKa = 6.86IQRR4 pKa = 11.84RR5 pKa = 11.84LLKK8 pKa = 9.02WLPVAMLAAFLLAGCSGDD26 pKa = 3.5NSSQGTPAGKK36 pKa = 10.02DD37 pKa = 2.97AVAQKK42 pKa = 9.94ATVTFKK48 pKa = 11.25VQFPEE53 pKa = 4.25SAVQKK58 pKa = 11.31AMIDD62 pKa = 3.56DD63 pKa = 3.9RR64 pKa = 11.84TVSVQVQWGDD74 pKa = 3.38YY75 pKa = 9.34YY76 pKa = 11.35NYY78 pKa = 10.96SFIDD82 pKa = 4.53SITLTPDD89 pKa = 2.8SSGMATATVTVPVGMLQFTAWANDD113 pKa = 3.52ANGMEE118 pKa = 4.92LEE120 pKa = 4.61MISTAGDD127 pKa = 3.06IVEE130 pKa = 4.76GNNTVYY136 pKa = 9.91LTFLGGDD143 pKa = 3.58WQFVDD148 pKa = 5.47ANDD151 pKa = 3.73APMPLSVGTGASSRR165 pKa = 11.84TLTGFSLGSAYY176 pKa = 10.81SHH178 pKa = 6.19GMYY181 pKa = 10.64AKK183 pKa = 10.64AKK185 pKa = 9.51VDD187 pKa = 3.36YY188 pKa = 7.62TKK190 pKa = 11.09PMGYY194 pKa = 10.34GDD196 pKa = 5.61YY197 pKa = 10.52RR198 pKa = 11.84LQWFDD203 pKa = 2.9TAGAVGSQMAAWADD217 pKa = 3.45NQLIGGVTNNTAFGSDD233 pKa = 3.8MLNLTTPANSDD244 pKa = 3.31NFSFWSFATAGDD256 pKa = 3.22RR257 pKa = 11.84VLFVVNSGPDD267 pKa = 2.84SGTIKK272 pKa = 10.82DD273 pKa = 3.82GAGNDD278 pKa = 3.4LTGAIDD284 pKa = 3.91AVADD288 pKa = 3.87AQILDD293 pKa = 3.72GTHH296 pKa = 6.46ISGHH300 pKa = 5.1LLEE303 pKa = 4.54MTFDD307 pKa = 3.83SVTGIQTPTRR317 pKa = 11.84TNIDD321 pKa = 3.55CAPYY325 pKa = 8.0WTYY328 pKa = 11.69AGAARR333 pKa = 11.84SAAIKK338 pKa = 10.29AAFASSSQGPAKK350 pKa = 10.18AAPGDD355 pKa = 4.07SSTVTATLVAAYY367 pKa = 8.01EE368 pKa = 4.25EE369 pKa = 5.5CNQQPYY375 pKa = 10.33QIDD378 pKa = 3.64GDD380 pKa = 3.96GDD382 pKa = 3.42QDD384 pKa = 3.33FWYY387 pKa = 10.87GDD389 pKa = 3.56YY390 pKa = 10.66LTFDD394 pKa = 4.0FNSNNRR400 pKa = 11.84YY401 pKa = 10.04DD402 pKa = 3.95PADD405 pKa = 3.79GDD407 pKa = 4.29TYY409 pKa = 11.48QDD411 pKa = 3.67TNNDD415 pKa = 3.18GNFDD419 pKa = 3.69FMIYY423 pKa = 10.29NQVDD427 pKa = 3.33GDD429 pKa = 4.07GDD431 pKa = 4.69GDD433 pKa = 4.14MLWDD437 pKa = 3.69YY438 pKa = 10.52MIVDD442 pKa = 3.8VNHH445 pKa = 6.58NGRR448 pKa = 11.84YY449 pKa = 9.21DD450 pKa = 3.46AADD453 pKa = 3.66GDD455 pKa = 4.7TYY457 pKa = 11.47QDD459 pKa = 3.3TDD461 pKa = 3.39SDD463 pKa = 4.09GKK465 pKa = 10.95FDD467 pKa = 4.45FVYY470 pKa = 10.55TPGDD474 pKa = 3.7LYY476 pKa = 11.21AITEE480 pKa = 4.27TFSNVVARR488 pKa = 11.84EE489 pKa = 3.63FRR491 pKa = 11.84ARR493 pKa = 11.84GSQIISQFTPSVLGTWGPAGYY514 pKa = 10.92QMTGQGFAVSFLDD527 pKa = 3.36ATHH530 pKa = 6.74YY531 pKa = 10.71IHH533 pKa = 7.66VEE535 pKa = 3.85DD536 pKa = 6.18GIPDD540 pKa = 3.78TDD542 pKa = 5.4PITGNIIGGPGMEE555 pKa = 4.21YY556 pKa = 9.23GTYY559 pKa = 10.33SVNATTGDD567 pKa = 3.61VTFTASKK574 pKa = 9.55DD575 pKa = 3.59TTGDD579 pKa = 2.88WGPAGTGTQVINFQFLDD596 pKa = 3.65NNHH599 pKa = 5.81FQITDD604 pKa = 3.74PVDD607 pKa = 3.34STTQTLGRR615 pKa = 11.84VTSNWNSAVGSWKK628 pKa = 10.42FGSEE632 pKa = 3.91ALGMQVVTLFEE643 pKa = 4.54GGNYY647 pKa = 9.51LVASSGPPGTSNLDD661 pKa = 3.33GMEE664 pKa = 4.12SGTYY668 pKa = 9.67SFQTISEE675 pKa = 4.69SIDD678 pKa = 3.13GTVTANITFNGMNSTIPDD696 pKa = 3.69LNDD699 pKa = 3.31TVIAAPGTSVTMPITMTNHH718 pKa = 5.7GNTIEE723 pKa = 4.13FDD725 pKa = 3.86NNGTPVAFSRR735 pKa = 11.84VQQ737 pKa = 2.84
MM1 pKa = 6.86IQRR4 pKa = 11.84RR5 pKa = 11.84LLKK8 pKa = 9.02WLPVAMLAAFLLAGCSGDD26 pKa = 3.5NSSQGTPAGKK36 pKa = 10.02DD37 pKa = 2.97AVAQKK42 pKa = 9.94ATVTFKK48 pKa = 11.25VQFPEE53 pKa = 4.25SAVQKK58 pKa = 11.31AMIDD62 pKa = 3.56DD63 pKa = 3.9RR64 pKa = 11.84TVSVQVQWGDD74 pKa = 3.38YY75 pKa = 9.34YY76 pKa = 11.35NYY78 pKa = 10.96SFIDD82 pKa = 4.53SITLTPDD89 pKa = 2.8SSGMATATVTVPVGMLQFTAWANDD113 pKa = 3.52ANGMEE118 pKa = 4.92LEE120 pKa = 4.61MISTAGDD127 pKa = 3.06IVEE130 pKa = 4.76GNNTVYY136 pKa = 9.91LTFLGGDD143 pKa = 3.58WQFVDD148 pKa = 5.47ANDD151 pKa = 3.73APMPLSVGTGASSRR165 pKa = 11.84TLTGFSLGSAYY176 pKa = 10.81SHH178 pKa = 6.19GMYY181 pKa = 10.64AKK183 pKa = 10.64AKK185 pKa = 9.51VDD187 pKa = 3.36YY188 pKa = 7.62TKK190 pKa = 11.09PMGYY194 pKa = 10.34GDD196 pKa = 5.61YY197 pKa = 10.52RR198 pKa = 11.84LQWFDD203 pKa = 2.9TAGAVGSQMAAWADD217 pKa = 3.45NQLIGGVTNNTAFGSDD233 pKa = 3.8MLNLTTPANSDD244 pKa = 3.31NFSFWSFATAGDD256 pKa = 3.22RR257 pKa = 11.84VLFVVNSGPDD267 pKa = 2.84SGTIKK272 pKa = 10.82DD273 pKa = 3.82GAGNDD278 pKa = 3.4LTGAIDD284 pKa = 3.91AVADD288 pKa = 3.87AQILDD293 pKa = 3.72GTHH296 pKa = 6.46ISGHH300 pKa = 5.1LLEE303 pKa = 4.54MTFDD307 pKa = 3.83SVTGIQTPTRR317 pKa = 11.84TNIDD321 pKa = 3.55CAPYY325 pKa = 8.0WTYY328 pKa = 11.69AGAARR333 pKa = 11.84SAAIKK338 pKa = 10.29AAFASSSQGPAKK350 pKa = 10.18AAPGDD355 pKa = 4.07SSTVTATLVAAYY367 pKa = 8.01EE368 pKa = 4.25EE369 pKa = 5.5CNQQPYY375 pKa = 10.33QIDD378 pKa = 3.64GDD380 pKa = 3.96GDD382 pKa = 3.42QDD384 pKa = 3.33FWYY387 pKa = 10.87GDD389 pKa = 3.56YY390 pKa = 10.66LTFDD394 pKa = 4.0FNSNNRR400 pKa = 11.84YY401 pKa = 10.04DD402 pKa = 3.95PADD405 pKa = 3.79GDD407 pKa = 4.29TYY409 pKa = 11.48QDD411 pKa = 3.67TNNDD415 pKa = 3.18GNFDD419 pKa = 3.69FMIYY423 pKa = 10.29NQVDD427 pKa = 3.33GDD429 pKa = 4.07GDD431 pKa = 4.69GDD433 pKa = 4.14MLWDD437 pKa = 3.69YY438 pKa = 10.52MIVDD442 pKa = 3.8VNHH445 pKa = 6.58NGRR448 pKa = 11.84YY449 pKa = 9.21DD450 pKa = 3.46AADD453 pKa = 3.66GDD455 pKa = 4.7TYY457 pKa = 11.47QDD459 pKa = 3.3TDD461 pKa = 3.39SDD463 pKa = 4.09GKK465 pKa = 10.95FDD467 pKa = 4.45FVYY470 pKa = 10.55TPGDD474 pKa = 3.7LYY476 pKa = 11.21AITEE480 pKa = 4.27TFSNVVARR488 pKa = 11.84EE489 pKa = 3.63FRR491 pKa = 11.84ARR493 pKa = 11.84GSQIISQFTPSVLGTWGPAGYY514 pKa = 10.92QMTGQGFAVSFLDD527 pKa = 3.36ATHH530 pKa = 6.74YY531 pKa = 10.71IHH533 pKa = 7.66VEE535 pKa = 3.85DD536 pKa = 6.18GIPDD540 pKa = 3.78TDD542 pKa = 5.4PITGNIIGGPGMEE555 pKa = 4.21YY556 pKa = 9.23GTYY559 pKa = 10.33SVNATTGDD567 pKa = 3.61VTFTASKK574 pKa = 9.55DD575 pKa = 3.59TTGDD579 pKa = 2.88WGPAGTGTQVINFQFLDD596 pKa = 3.65NNHH599 pKa = 5.81FQITDD604 pKa = 3.74PVDD607 pKa = 3.34STTQTLGRR615 pKa = 11.84VTSNWNSAVGSWKK628 pKa = 10.42FGSEE632 pKa = 3.91ALGMQVVTLFEE643 pKa = 4.54GGNYY647 pKa = 9.51LVASSGPPGTSNLDD661 pKa = 3.33GMEE664 pKa = 4.12SGTYY668 pKa = 9.67SFQTISEE675 pKa = 4.69SIDD678 pKa = 3.13GTVTANITFNGMNSTIPDD696 pKa = 3.69LNDD699 pKa = 3.31TVIAAPGTSVTMPITMTNHH718 pKa = 5.7GNTIEE723 pKa = 4.13FDD725 pKa = 3.86NNGTPVAFSRR735 pKa = 11.84VQQ737 pKa = 2.84
Molecular weight: 78.58 kDa
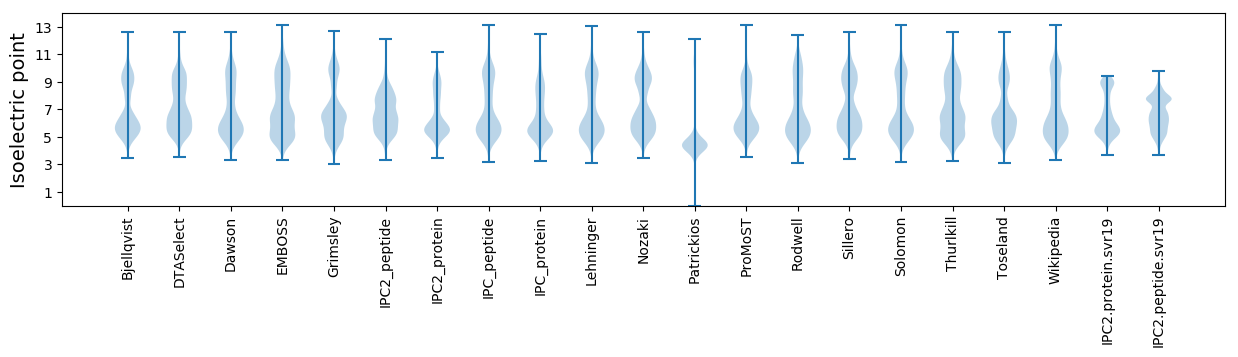
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A143BCB7|A0A143BCB7_9DELT Alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase OS=Desulfuromonas sp. DDH964 OX=1823759 GN=phnM PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.94RR4 pKa = 11.84TYY6 pKa = 9.71QPSRR10 pKa = 11.84VSRR13 pKa = 11.84KK14 pKa = 7.52RR15 pKa = 11.84THH17 pKa = 6.29GFRR20 pKa = 11.84KK21 pKa = 10.07RR22 pKa = 11.84MQSKK26 pKa = 10.01NGRR29 pKa = 11.84IVIKK33 pKa = 10.27RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.86GRR40 pKa = 11.84KK41 pKa = 8.29NLVVTIPSKK50 pKa = 10.99
MM1 pKa = 7.74SKK3 pKa = 8.94RR4 pKa = 11.84TYY6 pKa = 9.71QPSRR10 pKa = 11.84VSRR13 pKa = 11.84KK14 pKa = 7.52RR15 pKa = 11.84THH17 pKa = 6.29GFRR20 pKa = 11.84KK21 pKa = 10.07RR22 pKa = 11.84MQSKK26 pKa = 10.01NGRR29 pKa = 11.84IVIKK33 pKa = 10.27RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.86GRR40 pKa = 11.84KK41 pKa = 8.29NLVVTIPSKK50 pKa = 10.99
Molecular weight: 5.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1134986 |
29 |
3962 |
325.9 |
35.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.899 ± 0.06 | 1.208 ± 0.02 |
5.1 ± 0.027 | 6.425 ± 0.045 |
3.965 ± 0.03 | 8.41 ± 0.039 |
1.994 ± 0.021 | 5.107 ± 0.035 |
3.593 ± 0.039 | 11.844 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.022 ± 0.02 | 2.755 ± 0.031 |
5.053 ± 0.036 | 3.653 ± 0.024 |
7.091 ± 0.051 | 5.121 ± 0.031 |
4.989 ± 0.052 | 7.139 ± 0.037 |
1.135 ± 0.016 | 2.497 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
