
Beihai sobemo-like virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.61
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
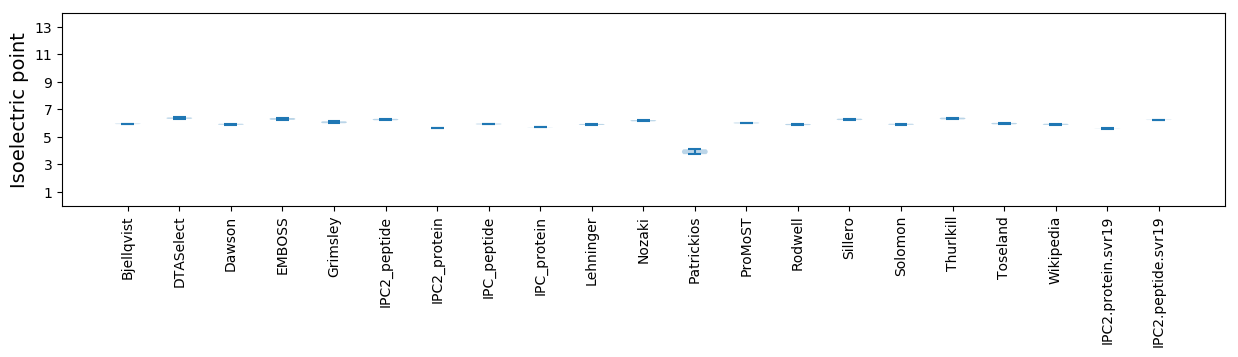
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEB3|A0A1L3KEB3_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 18 OX=1922689 PE=4 SV=1
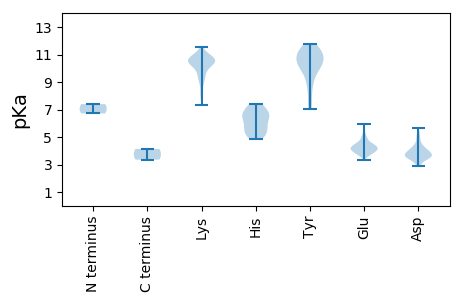
MM1 pKa = 7.39LKK3 pKa = 10.12ISGEE7 pKa = 3.92AADD10 pKa = 3.9QDD12 pKa = 3.79IEE14 pKa = 4.49VFDD17 pKa = 4.88ASPVMSRR24 pKa = 11.84MTPAQRR30 pKa = 11.84RR31 pKa = 11.84DD32 pKa = 3.56KK33 pKa = 10.54YY34 pKa = 11.72KK35 pKa = 10.75MMADD39 pKa = 3.81CEE41 pKa = 4.38DD42 pKa = 3.64VVVPMLQNVCSEE54 pKa = 4.56DD55 pKa = 3.58VPPDD59 pKa = 3.48YY60 pKa = 11.24LSFVAPNKK68 pKa = 9.85PLVDD72 pKa = 4.97EE73 pKa = 4.89IRR75 pKa = 11.84SQLGIEE81 pKa = 4.5FPLLTSDD88 pKa = 3.88EE89 pKa = 4.57SFGTLISTKK98 pKa = 9.33TSSGHH103 pKa = 5.0VLNPDD108 pKa = 3.46SSSGHH113 pKa = 5.01YY114 pKa = 7.02WAKK117 pKa = 10.47YY118 pKa = 7.58GANNGEE124 pKa = 4.15VFGWSPEE131 pKa = 3.71KK132 pKa = 10.7ARR134 pKa = 11.84YY135 pKa = 7.28TGEE138 pKa = 4.0NRR140 pKa = 11.84LSDD143 pKa = 3.96ANMIALFRR151 pKa = 11.84EE152 pKa = 4.52KK153 pKa = 10.37VVSRR157 pKa = 11.84MVEE160 pKa = 4.09LYY162 pKa = 10.25FMPVSDD168 pKa = 4.43VIKK171 pKa = 10.65VFIKK175 pKa = 10.63HH176 pKa = 5.9EE177 pKa = 3.83PHH179 pKa = 6.61KK180 pKa = 10.33PEE182 pKa = 4.58KK183 pKa = 10.25VKK185 pKa = 10.48QGKK188 pKa = 7.33WRR190 pKa = 11.84LIMGVCPTDD199 pKa = 3.36QICAHH204 pKa = 6.81LLFDD208 pKa = 3.67TVLAIFEE215 pKa = 4.93SIPLLSGTAIGWSPMMEE232 pKa = 4.21GGVSLYY238 pKa = 10.02RR239 pKa = 11.84AYY241 pKa = 11.02VGDD244 pKa = 4.16YY245 pKa = 11.64NMVTADD251 pKa = 3.68RR252 pKa = 11.84SSWDD256 pKa = 2.97WTVQFWLALTFISLMLYY273 pKa = 10.23LCNTRR278 pKa = 11.84SLLRR282 pKa = 11.84QRR284 pKa = 11.84IITNHH289 pKa = 5.69ILSMFGSRR297 pKa = 11.84KK298 pKa = 9.42FNVGASTFYY307 pKa = 9.88MHH309 pKa = 7.38ILGVLLSGWKK319 pKa = 8.16LTAFCNSLMQIICHH333 pKa = 6.2HH334 pKa = 6.58LACLAVGFQLPPPICMGDD352 pKa = 3.42DD353 pKa = 3.52TLQNDD358 pKa = 5.33PITYY362 pKa = 8.5MRR364 pKa = 11.84LFSNVLDD371 pKa = 4.6DD372 pKa = 4.2VDD374 pKa = 4.05PVAVDD379 pKa = 3.43RR380 pKa = 11.84EE381 pKa = 4.27YY382 pKa = 10.78WKK384 pKa = 10.78FLSSTGAILKK394 pKa = 9.2MVDD397 pKa = 2.87RR398 pKa = 11.84FEE400 pKa = 5.78DD401 pKa = 3.78YY402 pKa = 11.35SEE404 pKa = 4.28FCGFRR409 pKa = 11.84FIPGEE414 pKa = 4.33CYY416 pKa = 9.22PVYY419 pKa = 10.21DD420 pKa = 3.95VKK422 pKa = 11.12HH423 pKa = 5.8AVALHH428 pKa = 5.31NVPSEE433 pKa = 4.08FLHH436 pKa = 5.33STLRR440 pKa = 11.84SYY442 pKa = 11.36LWLYY446 pKa = 10.9CYY448 pKa = 10.72DD449 pKa = 3.65KK450 pKa = 11.05EE451 pKa = 4.75KK452 pKa = 11.53YY453 pKa = 8.49EE454 pKa = 4.3ALEE457 pKa = 3.61RR458 pKa = 11.84WLRR461 pKa = 11.84QTGGSTSFTRR471 pKa = 11.84LKK473 pKa = 10.42ILSQINGVAPPCIRR487 pKa = 11.84PLTWAA492 pKa = 4.13
MM1 pKa = 7.39LKK3 pKa = 10.12ISGEE7 pKa = 3.92AADD10 pKa = 3.9QDD12 pKa = 3.79IEE14 pKa = 4.49VFDD17 pKa = 4.88ASPVMSRR24 pKa = 11.84MTPAQRR30 pKa = 11.84RR31 pKa = 11.84DD32 pKa = 3.56KK33 pKa = 10.54YY34 pKa = 11.72KK35 pKa = 10.75MMADD39 pKa = 3.81CEE41 pKa = 4.38DD42 pKa = 3.64VVVPMLQNVCSEE54 pKa = 4.56DD55 pKa = 3.58VPPDD59 pKa = 3.48YY60 pKa = 11.24LSFVAPNKK68 pKa = 9.85PLVDD72 pKa = 4.97EE73 pKa = 4.89IRR75 pKa = 11.84SQLGIEE81 pKa = 4.5FPLLTSDD88 pKa = 3.88EE89 pKa = 4.57SFGTLISTKK98 pKa = 9.33TSSGHH103 pKa = 5.0VLNPDD108 pKa = 3.46SSSGHH113 pKa = 5.01YY114 pKa = 7.02WAKK117 pKa = 10.47YY118 pKa = 7.58GANNGEE124 pKa = 4.15VFGWSPEE131 pKa = 3.71KK132 pKa = 10.7ARR134 pKa = 11.84YY135 pKa = 7.28TGEE138 pKa = 4.0NRR140 pKa = 11.84LSDD143 pKa = 3.96ANMIALFRR151 pKa = 11.84EE152 pKa = 4.52KK153 pKa = 10.37VVSRR157 pKa = 11.84MVEE160 pKa = 4.09LYY162 pKa = 10.25FMPVSDD168 pKa = 4.43VIKK171 pKa = 10.65VFIKK175 pKa = 10.63HH176 pKa = 5.9EE177 pKa = 3.83PHH179 pKa = 6.61KK180 pKa = 10.33PEE182 pKa = 4.58KK183 pKa = 10.25VKK185 pKa = 10.48QGKK188 pKa = 7.33WRR190 pKa = 11.84LIMGVCPTDD199 pKa = 3.36QICAHH204 pKa = 6.81LLFDD208 pKa = 3.67TVLAIFEE215 pKa = 4.93SIPLLSGTAIGWSPMMEE232 pKa = 4.21GGVSLYY238 pKa = 10.02RR239 pKa = 11.84AYY241 pKa = 11.02VGDD244 pKa = 4.16YY245 pKa = 11.64NMVTADD251 pKa = 3.68RR252 pKa = 11.84SSWDD256 pKa = 2.97WTVQFWLALTFISLMLYY273 pKa = 10.23LCNTRR278 pKa = 11.84SLLRR282 pKa = 11.84QRR284 pKa = 11.84IITNHH289 pKa = 5.69ILSMFGSRR297 pKa = 11.84KK298 pKa = 9.42FNVGASTFYY307 pKa = 9.88MHH309 pKa = 7.38ILGVLLSGWKK319 pKa = 8.16LTAFCNSLMQIICHH333 pKa = 6.2HH334 pKa = 6.58LACLAVGFQLPPPICMGDD352 pKa = 3.42DD353 pKa = 3.52TLQNDD358 pKa = 5.33PITYY362 pKa = 8.5MRR364 pKa = 11.84LFSNVLDD371 pKa = 4.6DD372 pKa = 4.2VDD374 pKa = 4.05PVAVDD379 pKa = 3.43RR380 pKa = 11.84EE381 pKa = 4.27YY382 pKa = 10.78WKK384 pKa = 10.78FLSSTGAILKK394 pKa = 9.2MVDD397 pKa = 2.87RR398 pKa = 11.84FEE400 pKa = 5.78DD401 pKa = 3.78YY402 pKa = 11.35SEE404 pKa = 4.28FCGFRR409 pKa = 11.84FIPGEE414 pKa = 4.33CYY416 pKa = 9.22PVYY419 pKa = 10.21DD420 pKa = 3.95VKK422 pKa = 11.12HH423 pKa = 5.8AVALHH428 pKa = 5.31NVPSEE433 pKa = 4.08FLHH436 pKa = 5.33STLRR440 pKa = 11.84SYY442 pKa = 11.36LWLYY446 pKa = 10.9CYY448 pKa = 10.72DD449 pKa = 3.65KK450 pKa = 11.05EE451 pKa = 4.75KK452 pKa = 11.53YY453 pKa = 8.49EE454 pKa = 4.3ALEE457 pKa = 3.61RR458 pKa = 11.84WLRR461 pKa = 11.84QTGGSTSFTRR471 pKa = 11.84LKK473 pKa = 10.42ILSQINGVAPPCIRR487 pKa = 11.84PLTWAA492 pKa = 4.13
Molecular weight: 55.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEB3|A0A1L3KEB3_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 18 OX=1922689 PE=4 SV=1
MM1 pKa = 6.72NTSALFAEE9 pKa = 4.43FLEE12 pKa = 4.09YY13 pKa = 10.75RR14 pKa = 11.84KK15 pKa = 9.01MRR17 pKa = 11.84RR18 pKa = 11.84TMFQSSPLFEE28 pKa = 4.12WFEE31 pKa = 4.01LFIVKK36 pKa = 10.28AEE38 pKa = 4.11SLSRR42 pKa = 11.84MFDD45 pKa = 2.99IGKK48 pKa = 8.08MIPMALTVSSLSLSLCMMLHH68 pKa = 6.74GLALLLNVRR77 pKa = 11.84SRR79 pKa = 11.84ITSFLSSIFRR89 pKa = 11.84TLRR92 pKa = 11.84YY93 pKa = 9.86VLTRR97 pKa = 11.84LLPGVRR103 pKa = 11.84TYY105 pKa = 10.51LTVIALVVLCLGTLHH120 pKa = 7.3IALEE124 pKa = 4.36CFDD127 pKa = 4.3AFFVEE132 pKa = 5.54VPDD135 pKa = 3.85EE136 pKa = 4.16MEE138 pKa = 4.45PPVDD142 pKa = 3.34ISDD145 pKa = 3.58VSVATVSVLLICVGSYY161 pKa = 11.39ALLIWTNLDD170 pKa = 3.66RR171 pKa = 11.84LVLDD175 pKa = 3.46YY176 pKa = 11.28SYY178 pKa = 11.73VSSEE182 pKa = 3.82AAKK185 pKa = 10.7QIDD188 pKa = 4.4PNLMPGSVAMNDD200 pKa = 3.86PKK202 pKa = 10.81WLVTVGARR210 pKa = 11.84IGNMYY215 pKa = 9.25TIVGQAFQINQSYY228 pKa = 9.98VEE230 pKa = 4.04DD231 pKa = 4.8EE232 pKa = 3.72IWFITARR239 pKa = 11.84HH240 pKa = 4.87VVNHH244 pKa = 6.22IKK246 pKa = 10.55SYY248 pKa = 11.19DD249 pKa = 3.61NSIVTILTSTGKK261 pKa = 9.56PLTPIKK267 pKa = 10.08QVQLSDD273 pKa = 4.0SLPDD277 pKa = 3.55CMLLIVARR285 pKa = 11.84PKK287 pKa = 10.17GWNEE291 pKa = 3.66VGLKK295 pKa = 10.27VVDD298 pKa = 4.15PPFPSLSTDD307 pKa = 4.16FVCPVSIYY315 pKa = 10.97ARR317 pKa = 11.84DD318 pKa = 3.6VPEE321 pKa = 3.9QFQVRR326 pKa = 11.84ASRR329 pKa = 11.84GKK331 pKa = 10.31AGYY334 pKa = 8.98LVKK337 pKa = 10.58NAGMILHH344 pKa = 6.53HH345 pKa = 6.69TADD348 pKa = 4.37TVPGWSGSPILVTGDD363 pKa = 3.25EE364 pKa = 4.59GKK366 pKa = 10.81LEE368 pKa = 3.83VMGIHH373 pKa = 6.84LCCSGQTNNASRR385 pKa = 11.84NMGYY389 pKa = 11.02ALNAVLEE396 pKa = 4.75AYY398 pKa = 10.08VKK400 pKa = 9.42STYY403 pKa = 10.75GGVGNTVSQPVKK415 pKa = 10.61HH416 pKa = 6.02EE417 pKa = 4.19LVTEE421 pKa = 3.87ARR423 pKa = 11.84GRR425 pKa = 11.84PRR427 pKa = 11.84GRR429 pKa = 11.84PRR431 pKa = 11.84TRR433 pKa = 11.84RR434 pKa = 11.84VKK436 pKa = 10.61RR437 pKa = 11.84EE438 pKa = 3.91NKK440 pKa = 8.89SKK442 pKa = 10.79RR443 pKa = 11.84VFDD446 pKa = 3.7EE447 pKa = 4.32LYY449 pKa = 10.67NRR451 pKa = 11.84LEE453 pKa = 4.19SRR455 pKa = 11.84TGLQISRR462 pKa = 11.84IDD464 pKa = 3.71EE465 pKa = 4.09EE466 pKa = 4.53GANYY470 pKa = 10.66DD471 pKa = 4.16LFEE474 pKa = 4.42EE475 pKa = 5.12CSRR478 pKa = 11.84SLDD481 pKa = 3.4ATFVNHH487 pKa = 7.18HH488 pKa = 6.18EE489 pKa = 4.29VCQYY493 pKa = 10.53GCQSPEE499 pKa = 3.72EE500 pKa = 4.41CPLCKK505 pKa = 10.34NQVEE509 pKa = 4.52ILCGRR514 pKa = 11.84CDD516 pKa = 3.19WDD518 pKa = 3.86DD519 pKa = 4.37RR520 pKa = 11.84YY521 pKa = 11.39AHH523 pKa = 7.36DD524 pKa = 5.1EE525 pKa = 4.23DD526 pKa = 5.66CAMCNTLTAVCMCITTFGGQVAFANHH552 pKa = 5.56PTRR555 pKa = 11.84VDD557 pKa = 3.51EE558 pKa = 4.6NGMPEE563 pKa = 4.93VLLLWFGSDD572 pKa = 3.84YY573 pKa = 11.37YY574 pKa = 10.47PATTVPFSLNIGIDD588 pKa = 3.56PQPFILHH595 pKa = 5.29KK596 pKa = 9.78TIGGEE601 pKa = 3.95AKK603 pKa = 10.35GKK605 pKa = 10.34NKK607 pKa = 10.42GKK609 pKa = 10.56GFRR612 pKa = 11.84NLNKK616 pKa = 10.16YY617 pKa = 10.09AGSTSPSVSDD627 pKa = 3.54PRR629 pKa = 11.84SFLSWFDD636 pKa = 3.83DD637 pKa = 4.03PEE639 pKa = 5.92ADD641 pKa = 3.21RR642 pKa = 11.84TAFSFFPTVLPNKK655 pKa = 9.61DD656 pKa = 3.34VMWIVRR662 pKa = 11.84DD663 pKa = 3.87NNSGRR668 pKa = 11.84TSTWEE673 pKa = 3.52SDD675 pKa = 3.69YY676 pKa = 11.56LSDD679 pKa = 5.25QIIGRR684 pKa = 11.84LKK686 pKa = 8.39TWVNAEE692 pKa = 3.98AKK694 pKa = 10.71SPVLTKK700 pKa = 10.79AILDD704 pKa = 3.84EE705 pKa = 5.09DD706 pKa = 3.96DD707 pKa = 4.06QFIALHH713 pKa = 6.61DD714 pKa = 5.17SILEE718 pKa = 4.11DD719 pKa = 3.36EE720 pKa = 5.43FYY722 pKa = 10.82TVLKK726 pKa = 10.04VRR728 pKa = 11.84SVPISDD734 pKa = 3.78SLEE737 pKa = 4.1DD738 pKa = 4.1DD739 pKa = 4.45RR740 pKa = 11.84IAVEE744 pKa = 4.94KK745 pKa = 10.75ALEE748 pKa = 4.3DD749 pKa = 3.16YY750 pKa = 10.34RR751 pKa = 11.84YY752 pKa = 11.06AMEE755 pKa = 4.44QLGSTYY761 pKa = 10.98DD762 pKa = 3.61SLAMRR767 pKa = 11.84ARR769 pKa = 11.84LLIDD773 pKa = 4.82DD774 pKa = 4.56VSDD777 pKa = 3.88DD778 pKa = 3.9AAMISCQLAKK788 pKa = 10.42DD789 pKa = 3.47IQYY792 pKa = 10.54VCDD795 pKa = 4.15TISNLAQYY803 pKa = 9.61GAQLTQKK810 pKa = 10.81SNDD813 pKa = 2.99ICEE816 pKa = 4.38CSQVQISKK824 pKa = 10.19RR825 pKa = 11.84RR826 pKa = 11.84LMAKK830 pKa = 9.73KK831 pKa = 9.44RR832 pKa = 11.84AKK834 pKa = 9.74LRR836 pKa = 11.84KK837 pKa = 9.1EE838 pKa = 3.79VDD840 pKa = 4.08FQPSQAINDD849 pKa = 3.61IVSTCRR855 pKa = 11.84GTVNQALRR863 pKa = 11.84MAEE866 pKa = 4.3SIPLRR871 pKa = 11.84TYY873 pKa = 9.37RR874 pKa = 11.84TEE876 pKa = 4.22SYY878 pKa = 8.41TAQYY882 pKa = 10.2PGDD885 pKa = 4.41PPPNGPSSDD894 pKa = 3.72QPDD897 pKa = 3.82KK898 pKa = 10.98KK899 pKa = 10.69RR900 pKa = 11.84KK901 pKa = 8.03LTEE904 pKa = 3.72EE905 pKa = 3.91KK906 pKa = 10.63LRR908 pKa = 11.84AGFAKK913 pKa = 10.47ALNATYY919 pKa = 10.27DD920 pKa = 3.59YY921 pKa = 11.53GILTKK926 pKa = 10.88LKK928 pKa = 10.42DD929 pKa = 3.61FHH931 pKa = 7.06NEE933 pKa = 3.32PKK935 pKa = 10.37TFLDD939 pKa = 3.63FDD941 pKa = 4.2KK942 pKa = 11.2VRR944 pKa = 11.84QTTSEE949 pKa = 4.2KK950 pKa = 11.04EE951 pKa = 3.82EE952 pKa = 4.72DD953 pKa = 3.45SDD955 pKa = 4.12FSKK958 pKa = 10.88SASSRR963 pKa = 11.84LGSPTTPTEE972 pKa = 4.16SVTPRR977 pKa = 11.84QFHH980 pKa = 5.43PTLRR984 pKa = 11.84SFSRR988 pKa = 11.84TTLGGSTHH996 pKa = 6.43PHH998 pKa = 5.11QMMEE1002 pKa = 4.28VEE1004 pKa = 5.1NMAEE1008 pKa = 4.06RR1009 pKa = 11.84YY1010 pKa = 7.55GTLSNYY1016 pKa = 9.37IPQLPPRR1023 pKa = 11.84LQKK1026 pKa = 10.54EE1027 pKa = 4.19FQTLLNGKK1035 pKa = 9.01HH1036 pKa = 5.76SEE1038 pKa = 3.8EE1039 pKa = 4.0SRR1041 pKa = 11.84YY1042 pKa = 9.99LRR1044 pKa = 11.84PEE1046 pKa = 3.71GLMRR1050 pKa = 11.84SCPTKK1055 pKa = 10.98LEE1057 pKa = 4.18ADD1059 pKa = 3.92SLRR1062 pKa = 11.84GLLVSLKK1069 pKa = 10.77DD1070 pKa = 3.51NAYY1073 pKa = 10.51LSQEE1077 pKa = 3.98LMSEE1081 pKa = 4.18LMRR1084 pKa = 11.84RR1085 pKa = 11.84SEE1087 pKa = 3.98NN1088 pKa = 3.36
MM1 pKa = 6.72NTSALFAEE9 pKa = 4.43FLEE12 pKa = 4.09YY13 pKa = 10.75RR14 pKa = 11.84KK15 pKa = 9.01MRR17 pKa = 11.84RR18 pKa = 11.84TMFQSSPLFEE28 pKa = 4.12WFEE31 pKa = 4.01LFIVKK36 pKa = 10.28AEE38 pKa = 4.11SLSRR42 pKa = 11.84MFDD45 pKa = 2.99IGKK48 pKa = 8.08MIPMALTVSSLSLSLCMMLHH68 pKa = 6.74GLALLLNVRR77 pKa = 11.84SRR79 pKa = 11.84ITSFLSSIFRR89 pKa = 11.84TLRR92 pKa = 11.84YY93 pKa = 9.86VLTRR97 pKa = 11.84LLPGVRR103 pKa = 11.84TYY105 pKa = 10.51LTVIALVVLCLGTLHH120 pKa = 7.3IALEE124 pKa = 4.36CFDD127 pKa = 4.3AFFVEE132 pKa = 5.54VPDD135 pKa = 3.85EE136 pKa = 4.16MEE138 pKa = 4.45PPVDD142 pKa = 3.34ISDD145 pKa = 3.58VSVATVSVLLICVGSYY161 pKa = 11.39ALLIWTNLDD170 pKa = 3.66RR171 pKa = 11.84LVLDD175 pKa = 3.46YY176 pKa = 11.28SYY178 pKa = 11.73VSSEE182 pKa = 3.82AAKK185 pKa = 10.7QIDD188 pKa = 4.4PNLMPGSVAMNDD200 pKa = 3.86PKK202 pKa = 10.81WLVTVGARR210 pKa = 11.84IGNMYY215 pKa = 9.25TIVGQAFQINQSYY228 pKa = 9.98VEE230 pKa = 4.04DD231 pKa = 4.8EE232 pKa = 3.72IWFITARR239 pKa = 11.84HH240 pKa = 4.87VVNHH244 pKa = 6.22IKK246 pKa = 10.55SYY248 pKa = 11.19DD249 pKa = 3.61NSIVTILTSTGKK261 pKa = 9.56PLTPIKK267 pKa = 10.08QVQLSDD273 pKa = 4.0SLPDD277 pKa = 3.55CMLLIVARR285 pKa = 11.84PKK287 pKa = 10.17GWNEE291 pKa = 3.66VGLKK295 pKa = 10.27VVDD298 pKa = 4.15PPFPSLSTDD307 pKa = 4.16FVCPVSIYY315 pKa = 10.97ARR317 pKa = 11.84DD318 pKa = 3.6VPEE321 pKa = 3.9QFQVRR326 pKa = 11.84ASRR329 pKa = 11.84GKK331 pKa = 10.31AGYY334 pKa = 8.98LVKK337 pKa = 10.58NAGMILHH344 pKa = 6.53HH345 pKa = 6.69TADD348 pKa = 4.37TVPGWSGSPILVTGDD363 pKa = 3.25EE364 pKa = 4.59GKK366 pKa = 10.81LEE368 pKa = 3.83VMGIHH373 pKa = 6.84LCCSGQTNNASRR385 pKa = 11.84NMGYY389 pKa = 11.02ALNAVLEE396 pKa = 4.75AYY398 pKa = 10.08VKK400 pKa = 9.42STYY403 pKa = 10.75GGVGNTVSQPVKK415 pKa = 10.61HH416 pKa = 6.02EE417 pKa = 4.19LVTEE421 pKa = 3.87ARR423 pKa = 11.84GRR425 pKa = 11.84PRR427 pKa = 11.84GRR429 pKa = 11.84PRR431 pKa = 11.84TRR433 pKa = 11.84RR434 pKa = 11.84VKK436 pKa = 10.61RR437 pKa = 11.84EE438 pKa = 3.91NKK440 pKa = 8.89SKK442 pKa = 10.79RR443 pKa = 11.84VFDD446 pKa = 3.7EE447 pKa = 4.32LYY449 pKa = 10.67NRR451 pKa = 11.84LEE453 pKa = 4.19SRR455 pKa = 11.84TGLQISRR462 pKa = 11.84IDD464 pKa = 3.71EE465 pKa = 4.09EE466 pKa = 4.53GANYY470 pKa = 10.66DD471 pKa = 4.16LFEE474 pKa = 4.42EE475 pKa = 5.12CSRR478 pKa = 11.84SLDD481 pKa = 3.4ATFVNHH487 pKa = 7.18HH488 pKa = 6.18EE489 pKa = 4.29VCQYY493 pKa = 10.53GCQSPEE499 pKa = 3.72EE500 pKa = 4.41CPLCKK505 pKa = 10.34NQVEE509 pKa = 4.52ILCGRR514 pKa = 11.84CDD516 pKa = 3.19WDD518 pKa = 3.86DD519 pKa = 4.37RR520 pKa = 11.84YY521 pKa = 11.39AHH523 pKa = 7.36DD524 pKa = 5.1EE525 pKa = 4.23DD526 pKa = 5.66CAMCNTLTAVCMCITTFGGQVAFANHH552 pKa = 5.56PTRR555 pKa = 11.84VDD557 pKa = 3.51EE558 pKa = 4.6NGMPEE563 pKa = 4.93VLLLWFGSDD572 pKa = 3.84YY573 pKa = 11.37YY574 pKa = 10.47PATTVPFSLNIGIDD588 pKa = 3.56PQPFILHH595 pKa = 5.29KK596 pKa = 9.78TIGGEE601 pKa = 3.95AKK603 pKa = 10.35GKK605 pKa = 10.34NKK607 pKa = 10.42GKK609 pKa = 10.56GFRR612 pKa = 11.84NLNKK616 pKa = 10.16YY617 pKa = 10.09AGSTSPSVSDD627 pKa = 3.54PRR629 pKa = 11.84SFLSWFDD636 pKa = 3.83DD637 pKa = 4.03PEE639 pKa = 5.92ADD641 pKa = 3.21RR642 pKa = 11.84TAFSFFPTVLPNKK655 pKa = 9.61DD656 pKa = 3.34VMWIVRR662 pKa = 11.84DD663 pKa = 3.87NNSGRR668 pKa = 11.84TSTWEE673 pKa = 3.52SDD675 pKa = 3.69YY676 pKa = 11.56LSDD679 pKa = 5.25QIIGRR684 pKa = 11.84LKK686 pKa = 8.39TWVNAEE692 pKa = 3.98AKK694 pKa = 10.71SPVLTKK700 pKa = 10.79AILDD704 pKa = 3.84EE705 pKa = 5.09DD706 pKa = 3.96DD707 pKa = 4.06QFIALHH713 pKa = 6.61DD714 pKa = 5.17SILEE718 pKa = 4.11DD719 pKa = 3.36EE720 pKa = 5.43FYY722 pKa = 10.82TVLKK726 pKa = 10.04VRR728 pKa = 11.84SVPISDD734 pKa = 3.78SLEE737 pKa = 4.1DD738 pKa = 4.1DD739 pKa = 4.45RR740 pKa = 11.84IAVEE744 pKa = 4.94KK745 pKa = 10.75ALEE748 pKa = 4.3DD749 pKa = 3.16YY750 pKa = 10.34RR751 pKa = 11.84YY752 pKa = 11.06AMEE755 pKa = 4.44QLGSTYY761 pKa = 10.98DD762 pKa = 3.61SLAMRR767 pKa = 11.84ARR769 pKa = 11.84LLIDD773 pKa = 4.82DD774 pKa = 4.56VSDD777 pKa = 3.88DD778 pKa = 3.9AAMISCQLAKK788 pKa = 10.42DD789 pKa = 3.47IQYY792 pKa = 10.54VCDD795 pKa = 4.15TISNLAQYY803 pKa = 9.61GAQLTQKK810 pKa = 10.81SNDD813 pKa = 2.99ICEE816 pKa = 4.38CSQVQISKK824 pKa = 10.19RR825 pKa = 11.84RR826 pKa = 11.84LMAKK830 pKa = 9.73KK831 pKa = 9.44RR832 pKa = 11.84AKK834 pKa = 9.74LRR836 pKa = 11.84KK837 pKa = 9.1EE838 pKa = 3.79VDD840 pKa = 4.08FQPSQAINDD849 pKa = 3.61IVSTCRR855 pKa = 11.84GTVNQALRR863 pKa = 11.84MAEE866 pKa = 4.3SIPLRR871 pKa = 11.84TYY873 pKa = 9.37RR874 pKa = 11.84TEE876 pKa = 4.22SYY878 pKa = 8.41TAQYY882 pKa = 10.2PGDD885 pKa = 4.41PPPNGPSSDD894 pKa = 3.72QPDD897 pKa = 3.82KK898 pKa = 10.98KK899 pKa = 10.69RR900 pKa = 11.84KK901 pKa = 8.03LTEE904 pKa = 3.72EE905 pKa = 3.91KK906 pKa = 10.63LRR908 pKa = 11.84AGFAKK913 pKa = 10.47ALNATYY919 pKa = 10.27DD920 pKa = 3.59YY921 pKa = 11.53GILTKK926 pKa = 10.88LKK928 pKa = 10.42DD929 pKa = 3.61FHH931 pKa = 7.06NEE933 pKa = 3.32PKK935 pKa = 10.37TFLDD939 pKa = 3.63FDD941 pKa = 4.2KK942 pKa = 11.2VRR944 pKa = 11.84QTTSEE949 pKa = 4.2KK950 pKa = 11.04EE951 pKa = 3.82EE952 pKa = 4.72DD953 pKa = 3.45SDD955 pKa = 4.12FSKK958 pKa = 10.88SASSRR963 pKa = 11.84LGSPTTPTEE972 pKa = 4.16SVTPRR977 pKa = 11.84QFHH980 pKa = 5.43PTLRR984 pKa = 11.84SFSRR988 pKa = 11.84TTLGGSTHH996 pKa = 6.43PHH998 pKa = 5.11QMMEE1002 pKa = 4.28VEE1004 pKa = 5.1NMAEE1008 pKa = 4.06RR1009 pKa = 11.84YY1010 pKa = 7.55GTLSNYY1016 pKa = 9.37IPQLPPRR1023 pKa = 11.84LQKK1026 pKa = 10.54EE1027 pKa = 4.19FQTLLNGKK1035 pKa = 9.01HH1036 pKa = 5.76SEE1038 pKa = 3.8EE1039 pKa = 4.0SRR1041 pKa = 11.84YY1042 pKa = 9.99LRR1044 pKa = 11.84PEE1046 pKa = 3.71GLMRR1050 pKa = 11.84SCPTKK1055 pKa = 10.98LEE1057 pKa = 4.18ADD1059 pKa = 3.92SLRR1062 pKa = 11.84GLLVSLKK1069 pKa = 10.77DD1070 pKa = 3.51NAYY1073 pKa = 10.51LSQEE1077 pKa = 3.98LMSEE1081 pKa = 4.18LMRR1084 pKa = 11.84RR1085 pKa = 11.84SEE1087 pKa = 3.98NN1088 pKa = 3.36
Molecular weight: 122.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1580 |
492 |
1088 |
790.0 |
89.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.013 ± 0.167 | 2.405 ± 0.123 |
6.139 ± 0.127 | 5.696 ± 0.424 |
4.241 ± 0.436 | 5.443 ± 0.129 |
1.962 ± 0.247 | 5.0 ± 0.253 |
4.747 ± 0.143 | 10.127 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.228 ± 0.434 | 3.797 ± 0.283 |
5.316 ± 0.089 | 3.291 ± 0.336 |
5.823 ± 0.49 | 8.608 ± 0.142 |
5.949 ± 0.556 | 7.089 ± 0.224 |
1.519 ± 0.477 | 3.608 ± 0.237 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
