
Persicimonas caeni
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Bradymonadales; Bradymonadaceae; Persicimonas
Average proteome isoelectric point is 5.41
Get precalculated fractions of proteins
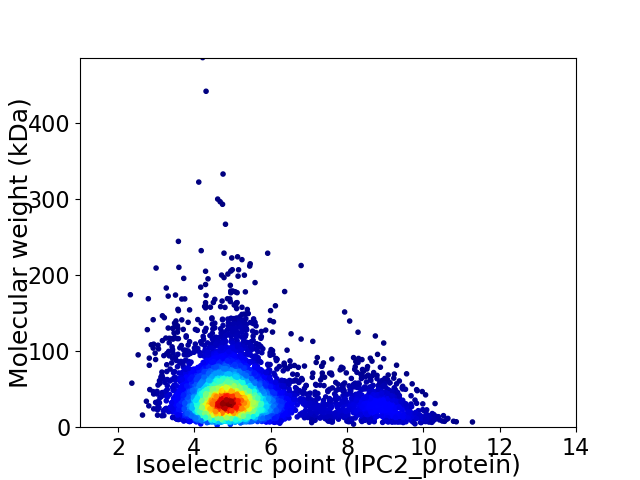
Virtual 2D-PAGE plot for 5752 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y6Q1Y0|A0A4Y6Q1Y0_9DELT FHA domain-containing protein OS=Persicimonas caeni OX=2292766 GN=FIV42_27950 PE=3 SV=1
MM1 pKa = 8.04DD2 pKa = 4.09VMEE5 pKa = 4.77KK6 pKa = 10.23CAAVRR11 pKa = 11.84PSASGLVRR19 pKa = 11.84RR20 pKa = 11.84FRR22 pKa = 11.84SLVALSAFVAAASWTVDD39 pKa = 2.91ASASCTTIEE48 pKa = 4.61DD49 pKa = 4.74FEE51 pKa = 5.27SGWPNSPWTVYY62 pKa = 10.57DD63 pKa = 3.7LPGTRR68 pKa = 11.84TTTYY72 pKa = 10.43SRR74 pKa = 11.84SGSYY78 pKa = 10.28GVQNPDD84 pKa = 2.05WVYY87 pKa = 9.77RR88 pKa = 11.84TDD90 pKa = 3.35VTIGQQAGEE99 pKa = 4.1RR100 pKa = 11.84LGLWFHH106 pKa = 6.89GSTASSGRR114 pKa = 11.84VYY116 pKa = 10.84LGFDD120 pKa = 3.69ADD122 pKa = 3.69GSGAKK127 pKa = 9.26TFIASLNTTDD137 pKa = 5.97IRR139 pKa = 11.84FQDD142 pKa = 3.4NAGYY146 pKa = 10.18NHH148 pKa = 6.43TQLNQTSQTFNSQWYY163 pKa = 7.77YY164 pKa = 11.56LEE166 pKa = 4.79VEE168 pKa = 4.49FNGGGSVTGRR178 pKa = 11.84LYY180 pKa = 11.07DD181 pKa = 3.5SDD183 pKa = 3.82GTTLINSLTQSFSGSVVGGVVLRR206 pKa = 11.84AFDD209 pKa = 4.78DD210 pKa = 3.83NVIDD214 pKa = 5.6DD215 pKa = 5.26VIHH218 pKa = 5.91CTPNDD223 pKa = 3.57APTASNDD230 pKa = 3.44SYY232 pKa = 11.95GVDD235 pKa = 3.56EE236 pKa = 6.3DD237 pKa = 4.35GLLDD241 pKa = 3.65VSPSGVLANDD251 pKa = 3.59TDD253 pKa = 4.38PDD255 pKa = 3.94GDD257 pKa = 4.13ALTASVVSGPSNGTLNLSANGGFTYY282 pKa = 10.56EE283 pKa = 4.22PAPNFHH289 pKa = 6.79GSDD292 pKa = 3.11SFTYY296 pKa = 10.12RR297 pKa = 11.84ASDD300 pKa = 3.5GDD302 pKa = 4.07GGSDD306 pKa = 3.03TATVNISVGSVNDD319 pKa = 3.66APVFTSTPVTSATEE333 pKa = 3.93DD334 pKa = 3.2QPYY337 pKa = 10.46SYY339 pKa = 10.78LVTATDD345 pKa = 3.23VDD347 pKa = 3.99TGDD350 pKa = 3.88SLTISLATRR359 pKa = 11.84PSWLGLSLGSGGSATLSGTPSNADD383 pKa = 3.29VGPHH387 pKa = 4.61SVEE390 pKa = 3.96VVVRR394 pKa = 11.84DD395 pKa = 3.83GNGGSDD401 pKa = 3.31TQTFTIDD408 pKa = 3.34VQNVNDD414 pKa = 3.88APYY417 pKa = 10.15FVDD420 pKa = 3.98PTPQDD425 pKa = 3.27GATLSVVEE433 pKa = 4.67GDD435 pKa = 3.66TLSFTLAGADD445 pKa = 4.32DD446 pKa = 5.46DD447 pKa = 6.03GDD449 pKa = 3.89TLTYY453 pKa = 10.7GVDD456 pKa = 3.69SVPTGASFDD465 pKa = 3.71ASTGAFSWTPTWW477 pKa = 3.35
MM1 pKa = 8.04DD2 pKa = 4.09VMEE5 pKa = 4.77KK6 pKa = 10.23CAAVRR11 pKa = 11.84PSASGLVRR19 pKa = 11.84RR20 pKa = 11.84FRR22 pKa = 11.84SLVALSAFVAAASWTVDD39 pKa = 2.91ASASCTTIEE48 pKa = 4.61DD49 pKa = 4.74FEE51 pKa = 5.27SGWPNSPWTVYY62 pKa = 10.57DD63 pKa = 3.7LPGTRR68 pKa = 11.84TTTYY72 pKa = 10.43SRR74 pKa = 11.84SGSYY78 pKa = 10.28GVQNPDD84 pKa = 2.05WVYY87 pKa = 9.77RR88 pKa = 11.84TDD90 pKa = 3.35VTIGQQAGEE99 pKa = 4.1RR100 pKa = 11.84LGLWFHH106 pKa = 6.89GSTASSGRR114 pKa = 11.84VYY116 pKa = 10.84LGFDD120 pKa = 3.69ADD122 pKa = 3.69GSGAKK127 pKa = 9.26TFIASLNTTDD137 pKa = 5.97IRR139 pKa = 11.84FQDD142 pKa = 3.4NAGYY146 pKa = 10.18NHH148 pKa = 6.43TQLNQTSQTFNSQWYY163 pKa = 7.77YY164 pKa = 11.56LEE166 pKa = 4.79VEE168 pKa = 4.49FNGGGSVTGRR178 pKa = 11.84LYY180 pKa = 11.07DD181 pKa = 3.5SDD183 pKa = 3.82GTTLINSLTQSFSGSVVGGVVLRR206 pKa = 11.84AFDD209 pKa = 4.78DD210 pKa = 3.83NVIDD214 pKa = 5.6DD215 pKa = 5.26VIHH218 pKa = 5.91CTPNDD223 pKa = 3.57APTASNDD230 pKa = 3.44SYY232 pKa = 11.95GVDD235 pKa = 3.56EE236 pKa = 6.3DD237 pKa = 4.35GLLDD241 pKa = 3.65VSPSGVLANDD251 pKa = 3.59TDD253 pKa = 4.38PDD255 pKa = 3.94GDD257 pKa = 4.13ALTASVVSGPSNGTLNLSANGGFTYY282 pKa = 10.56EE283 pKa = 4.22PAPNFHH289 pKa = 6.79GSDD292 pKa = 3.11SFTYY296 pKa = 10.12RR297 pKa = 11.84ASDD300 pKa = 3.5GDD302 pKa = 4.07GGSDD306 pKa = 3.03TATVNISVGSVNDD319 pKa = 3.66APVFTSTPVTSATEE333 pKa = 3.93DD334 pKa = 3.2QPYY337 pKa = 10.46SYY339 pKa = 10.78LVTATDD345 pKa = 3.23VDD347 pKa = 3.99TGDD350 pKa = 3.88SLTISLATRR359 pKa = 11.84PSWLGLSLGSGGSATLSGTPSNADD383 pKa = 3.29VGPHH387 pKa = 4.61SVEE390 pKa = 3.96VVVRR394 pKa = 11.84DD395 pKa = 3.83GNGGSDD401 pKa = 3.31TQTFTIDD408 pKa = 3.34VQNVNDD414 pKa = 3.88APYY417 pKa = 10.15FVDD420 pKa = 3.98PTPQDD425 pKa = 3.27GATLSVVEE433 pKa = 4.67GDD435 pKa = 3.66TLSFTLAGADD445 pKa = 4.32DD446 pKa = 5.46DD447 pKa = 6.03GDD449 pKa = 3.89TLTYY453 pKa = 10.7GVDD456 pKa = 3.69SVPTGASFDD465 pKa = 3.71ASTGAFSWTPTWW477 pKa = 3.35
Molecular weight: 49.72 kDa
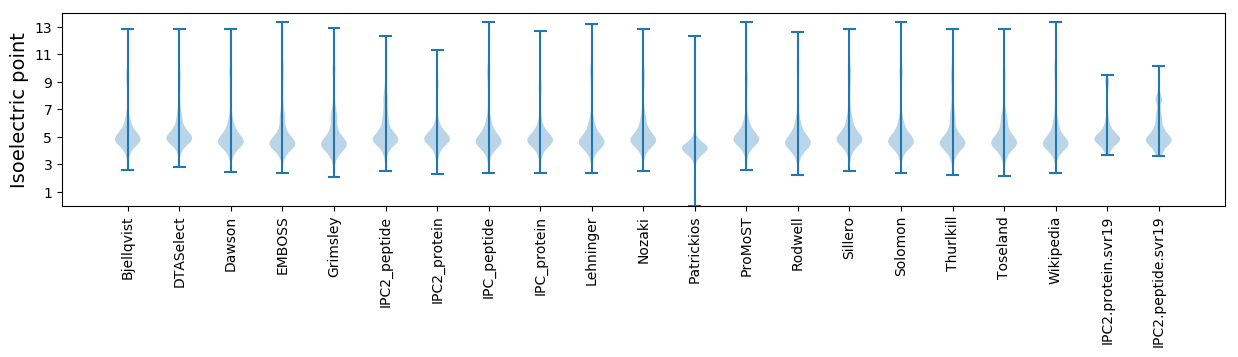
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y6PTH9|A0A4Y6PTH9_9DELT Glycosyltransferase family 2 protein OS=Persicimonas caeni OX=2292766 GN=FIV42_13020 PE=4 SV=1
MM1 pKa = 8.03PKK3 pKa = 8.96RR4 pKa = 11.84TYY6 pKa = 10.02QPSRR10 pKa = 11.84KK11 pKa = 9.22KK12 pKa = 10.07RR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 6.37THH17 pKa = 5.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MKK24 pKa = 10.52SKK26 pKa = 10.66GGRR29 pKa = 11.84KK30 pKa = 5.41TLKK33 pKa = 9.62RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.47GRR40 pKa = 11.84KK41 pKa = 8.27RR42 pKa = 11.84LSPRR46 pKa = 11.84KK47 pKa = 7.85FHH49 pKa = 6.62KK50 pKa = 9.47RR51 pKa = 11.84TRR53 pKa = 11.84RR54 pKa = 11.84TT55 pKa = 3.18
MM1 pKa = 8.03PKK3 pKa = 8.96RR4 pKa = 11.84TYY6 pKa = 10.02QPSRR10 pKa = 11.84KK11 pKa = 9.22KK12 pKa = 10.07RR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 6.37THH17 pKa = 5.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MKK24 pKa = 10.52SKK26 pKa = 10.66GGRR29 pKa = 11.84KK30 pKa = 5.41TLKK33 pKa = 9.62RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.47GRR40 pKa = 11.84KK41 pKa = 8.27RR42 pKa = 11.84LSPRR46 pKa = 11.84KK47 pKa = 7.85FHH49 pKa = 6.62KK50 pKa = 9.47RR51 pKa = 11.84TRR53 pKa = 11.84RR54 pKa = 11.84TT55 pKa = 3.18
Molecular weight: 6.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2306908 |
23 |
4216 |
401.1 |
44.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.287 ± 0.029 | 1.246 ± 0.026 |
7.343 ± 0.046 | 7.791 ± 0.044 |
3.601 ± 0.018 | 8.131 ± 0.034 |
2.112 ± 0.015 | 4.208 ± 0.021 |
3.373 ± 0.034 | 9.456 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.06 ± 0.014 | 2.71 ± 0.02 |
4.859 ± 0.021 | 3.521 ± 0.022 |
6.666 ± 0.04 | 5.749 ± 0.026 |
5.411 ± 0.035 | 7.566 ± 0.028 |
1.378 ± 0.015 | 2.532 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
